Project
DANIO-CODE
Navigation
Downloads
Results for hoxd4a
Z-value: 0.60
Transcription factors associated with hoxd4a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxd4a
|
ENSDARG00000059276 | homeobox D4a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxd4a | dr10_dc_chr9_-_1949904_1949953 | -0.39 | 1.4e-01 | Click! |
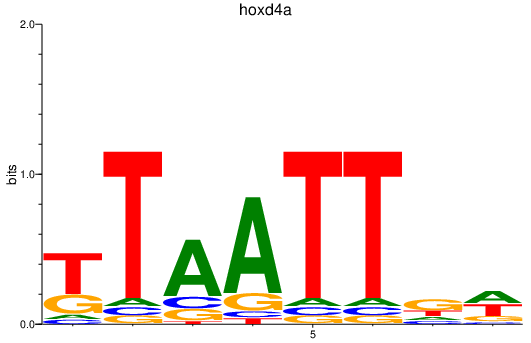
Activity profile of hoxd4a motif
Sorted Z-values of hoxd4a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxd4a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_26240772 | 1.30 |
ENSDART00000131394
|
fhdc3
|
FH2 domain containing 3 |
| chr24_-_9844547 | 1.24 |
|
|
|
| chr17_-_2401636 | 1.15 |
ENSDART00000013506
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr10_-_25246786 | 1.09 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr24_+_1110206 | 0.96 |
ENSDART00000152063
|
BX005461.1
|
ENSDARG00000096447 |
| chr7_+_16771283 | 0.90 |
ENSDART00000171275
|
CU929054.1
|
ENSDARG00000102605 |
| chr3_+_42383724 | 0.86 |
|
|
|
| chr6_+_18941186 | 0.84 |
ENSDART00000044519
|
cx44.2
|
connexin 44.2 |
| chr1_-_18118467 | 0.83 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr1_-_44628278 | 0.80 |
ENSDART00000133572
|
si:ch73-90k17.1
|
si:ch73-90k17.1 |
| chr19_-_44161905 | 0.73 |
ENSDART00000058491
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr16_+_29574449 | 0.73 |
ENSDART00000148450
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr22_+_25754857 | 0.67 |
ENSDART00000174421
|
AL929192.1
|
ENSDARG00000106684 |
| chr24_-_9838947 | 0.67 |
ENSDART00000138576
|
zgc:171977
|
zgc:171977 |
| chr7_+_44463158 | 0.66 |
ENSDART00000066380
|
ca7
|
carbonic anhydrase VII |
| chr17_-_45021393 | 0.64 |
|
|
|
| chr1_+_43471879 | 0.64 |
ENSDART00000166324
|
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr7_-_20330881 | 0.61 |
ENSDART00000169750
|
si:dkey-19b23.11
|
si:dkey-19b23.11 |
| chr11_-_18078147 | 0.60 |
ENSDART00000177625
|
zgc:173545
|
zgc:173545 |
| chr19_-_44161951 | 0.57 |
ENSDART00000058491
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr12_+_20222000 | 0.57 |
ENSDART00000150020
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr14_-_31277840 | 0.56 |
ENSDART00000016592
|
mmgt1
|
membrane magnesium transporter 1 |
| chr20_+_6545194 | 0.55 |
ENSDART00000159829
|
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr15_-_23786063 | 0.55 |
ENSDART00000109318
|
zc3h4
|
zinc finger CCCH-type containing 4 |
| chr4_-_14193396 | 0.52 |
ENSDART00000101812
ENSDART00000143804 |
pus7l
|
pseudouridylate synthase 7-like |
| chr14_-_33141111 | 0.51 |
ENSDART00000147059
|
lamp2
|
lysosomal-associated membrane protein 2 |
| chr8_+_45326435 | 0.51 |
ENSDART00000134161
|
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr6_+_18941289 | 0.50 |
ENSDART00000044519
|
cx44.2
|
connexin 44.2 |
| chr23_-_24416911 | 0.47 |
ENSDART00000166392
|
fam131c
|
family with sequence similarity 131, member C |
| chr17_+_10582044 | 0.46 |
ENSDART00000051527
|
tbpl2
|
TATA box binding protein like 2 |
| chr22_+_25755073 | 0.46 |
ENSDART00000174421
|
AL929192.1
|
ENSDARG00000106684 |
| chr20_+_7337986 | 0.46 |
ENSDART00000140222
|
dsg2.1
|
desmoglein 2, tandem duplicate 1 |
| chr20_-_46079578 | 0.46 |
ENSDART00000153228
|
AL929237.3
|
ENSDARG00000096676 |
| chr10_-_28494204 | 0.46 |
ENSDART00000131220
|
btg3
|
B-cell translocation gene 3 |
| chr9_-_22014861 | 0.44 |
ENSDART00000101986
|
mrpl30
|
mitochondrial ribosomal protein L30 |
| chr16_+_35448634 | 0.44 |
ENSDART00000171608
|
rab42b
|
RAB42, member RAS oncogene family |
| chr2_+_56534374 | 0.44 |
ENSDART00000113964
|
POLR2E (1 of many)
|
RNA polymerase II subunit E |
| chr14_-_33138710 | 0.43 |
ENSDART00000132813
|
lamp2
|
lysosomal-associated membrane protein 2 |
| chr3_-_32205961 | 0.43 |
ENSDART00000156551
|
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr6_+_18941135 | 0.42 |
ENSDART00000044519
|
cx44.2
|
connexin 44.2 |
| chr7_+_5835252 | 0.42 |
ENSDART00000170763
|
HIST2H2AB (1 of many)
|
histone cluster 2 H2A family member b |
| chr24_-_41275160 | 0.39 |
|
|
|
| chr10_-_26240909 | 0.39 |
ENSDART00000131394
|
fhdc3
|
FH2 domain containing 3 |
| chr6_-_30702168 | 0.39 |
ENSDART00000065211
|
pars2
|
prolyl-tRNA synthetase 2, mitochondrial (putative) |
| chr1_+_18118735 | 0.39 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr5_-_15971338 | 0.38 |
ENSDART00000110437
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr14_-_880799 | 0.38 |
ENSDART00000031992
|
rgs14a
|
regulator of G protein signaling 14a |
| chr15_-_23849212 | 0.36 |
ENSDART00000059354
|
rad1
|
RAD1 homolog (S. pombe) |
| chr10_-_36848872 | 0.35 |
|
|
|
| chr5_+_32687626 | 0.35 |
ENSDART00000146759
|
med22
|
mediator complex subunit 22 |
| chr3_+_30187036 | 0.34 |
ENSDART00000151006
|
CR936968.1
|
ENSDARG00000096295 |
| chr17_-_25630635 | 0.34 |
ENSDART00000149060
|
ppp1cb
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr11_+_11319810 | 0.34 |
ENSDART00000162486
|
si:dkey-23f9.4
|
si:dkey-23f9.4 |
| chr24_+_39339391 | 0.34 |
ENSDART00000168705
|
si:ch73-103b11.2
|
si:ch73-103b11.2 |
| chr21_-_19883182 | 0.33 |
ENSDART00000065670
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr22_-_10026610 | 0.32 |
ENSDART00000179409
|
si:ch211-222k6.1
|
si:ch211-222k6.1 |
| chr3_-_32741894 | 0.30 |
ENSDART00000075277
|
zgc:113090
|
zgc:113090 |
| chr3_+_30927580 | 0.30 |
ENSDART00000153381
|
si:dkey-66i24.7
|
si:dkey-66i24.7 |
| chr16_-_2632781 | 0.29 |
ENSDART00000148543
|
cdcp1a
|
CUB domain containing protein 1a |
| chr1_-_44513922 | 0.29 |
ENSDART00000014727
|
ddx39aa
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39Aa |
| chr22_-_38947928 | 0.29 |
ENSDART00000008365
|
ncbp2
|
nuclear cap binding protein subunit 2 |
| chr10_-_36848930 | 0.29 |
|
|
|
| chr5_+_20418001 | 0.29 |
ENSDART00000131838
|
si:dkey-174n20.1
|
si:dkey-174n20.1 |
| chr10_-_18535079 | 0.29 |
|
|
|
| chr10_-_25808063 | 0.29 |
ENSDART00000134176
|
postna
|
periostin, osteoblast specific factor a |
| chr22_+_11943005 | 0.28 |
ENSDART00000105788
|
mgat5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr18_+_13235539 | 0.28 |
ENSDART00000032151
|
cotl1
|
coactosin-like F-actin binding protein 1 |
| chr9_-_22014768 | 0.28 |
ENSDART00000101986
|
mrpl30
|
mitochondrial ribosomal protein L30 |
| chr20_-_4115423 | 0.28 |
ENSDART00000064365
|
si:ch73-111k22.2
|
si:ch73-111k22.2 |
| chr4_+_5823740 | 0.28 |
ENSDART00000121743
|
si:ch73-352p4.5
|
si:ch73-352p4.5 |
| chr11_+_42352106 | 0.27 |
ENSDART00000169938
|
il17rd
|
interleukin 17 receptor D |
| chr4_-_29714229 | 0.27 |
|
|
|
| chr20_-_3220106 | 0.27 |
ENSDART00000123331
|
spint1b
|
serine peptidase inhibitor, Kunitz type 1 b |
| chr15_-_16936518 | 0.26 |
ENSDART00000142870
ENSDART00000171857 ENSDART00000152633 |
tyw1
|
tRNA-yW synthesizing protein 1 homolog (S. cerevisiae) |
| chr15_-_3023850 | 0.26 |
|
|
|
| chr14_-_31149030 | 0.26 |
ENSDART00000018347
|
cab39l1
|
calcium binding protein 39, like 1 |
| chr23_-_19125310 | 0.25 |
ENSDART00000140866
|
arfgef2
|
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) |
| chr17_+_26785230 | 0.25 |
ENSDART00000023470
|
pgrmc2
|
progesterone receptor membrane component 2 |
| chr20_+_36725342 | 0.25 |
ENSDART00000161241
|
ephx1
|
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr16_-_13723778 | 0.25 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr16_-_13790460 | 0.25 |
ENSDART00000047452
|
ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr7_-_6297068 | 0.24 |
ENSDART00000172954
|
si:ch1073-153i20.4
|
si:ch1073-153i20.4 |
| chr16_+_22114192 | 0.24 |
ENSDART00000167919
|
znf687a
|
zinc finger protein 687a |
| chr18_+_3475225 | 0.24 |
ENSDART00000169814
|
serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chr25_-_5805538 | 0.24 |
|
|
|
| chr2_-_45282536 | 0.23 |
ENSDART00000123916
|
eif4e2
|
eukaryotic translation initiation factor 4E family member 2 |
| chr20_+_31375136 | 0.22 |
|
|
|
| chr7_-_41232765 | 0.22 |
ENSDART00000173577
|
ENSDARG00000105669
|
ENSDARG00000105669 |
| chr10_-_36747849 | 0.22 |
ENSDART00000122375
|
mrpl48
|
mitochondrial ribosomal protein L48 |
| chr13_+_11307037 | 0.22 |
ENSDART00000138312
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr16_-_10946309 | 0.22 |
ENSDART00000049148
|
rabac1
|
Rab acceptor 1 (prenylated) |
| chr13_-_49294664 | 0.21 |
ENSDART00000165868
|
tomm20a
|
translocase of outer mitochondrial membrane 20 |
| chr12_-_7412438 | 0.21 |
|
|
|
| chr2_+_9957535 | 0.21 |
ENSDART00000018524
|
pcyt1aa
|
phosphate cytidylyltransferase 1, choline, alpha a |
| chr18_-_19016801 | 0.21 |
ENSDART00000129776
|
vwa9
|
von Willebrand factor A domain containing 9 |
| chr19_+_17449569 | 0.21 |
ENSDART00000178325
|
SCARNA16
|
Small Cajal body specific RNA 16 |
| chr15_+_47633835 | 0.21 |
|
|
|
| chr5_+_6391432 | 0.20 |
ENSDART00000170564
ENSDART00000086666 |
stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr23_+_24685148 | 0.20 |
ENSDART00000134978
|
nckap5l
|
NCK-associated protein 5-like |
| chr5_+_4906690 | 0.20 |
ENSDART00000124022
|
mapkap1
|
mitogen-activated protein kinase associated protein 1 |
| chr9_+_38648006 | 0.19 |
ENSDART00000142512
|
mcm3ap
|
minichromosome maintenance complex component 3 associated protein |
| chr12_-_29118423 | 0.19 |
ENSDART00000153175
|
si:ch211-214e3.5
|
si:ch211-214e3.5 |
| chr7_+_13238684 | 0.19 |
ENSDART00000053535
|
arih1l
|
ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 like |
| chr17_-_31694979 | 0.19 |
ENSDART00000077307
|
pomt2
|
protein-O-mannosyltransferase 2 |
| chr8_+_23084130 | 0.19 |
ENSDART00000025171
|
ythdf1
|
YTH N(6)-methyladenosine RNA binding protein 1 |
| chr9_+_29806055 | 0.18 |
ENSDART00000143493
|
phf11
|
PHD finger protein 11 |
| chr1_+_518777 | 0.18 |
ENSDART00000109083
|
txnl4b
|
thioredoxin-like 4B |
| chr17_-_11285631 | 0.18 |
ENSDART00000130105
|
psma3
|
proteasome subunit alpha 3 |
| chr11_-_19909409 | 0.18 |
ENSDART00000121722
|
si:dkey-274m17.3
|
si:dkey-274m17.3 |
| chr6_-_44163685 | 0.18 |
ENSDART00000035513
|
shq1
|
SHQ1, H/ACA ribonucleoprotein assembly factor |
| chr20_-_16271738 | 0.17 |
ENSDART00000012476
|
coa7
|
cytochrome c oxidase assembly factor 7 |
| chr5_+_24378828 | 0.17 |
ENSDART00000111302
|
rhbdd3
|
rhomboid domain containing 3 |
| chr13_+_36459625 | 0.17 |
ENSDART00000030211
|
gmfb
|
glia maturation factor, beta |
| chr17_-_28780394 | 0.17 |
ENSDART00000167897
ENSDART00000134735 |
scfd1
|
sec1 family domain containing 1 |
| chr24_-_36248243 | 0.16 |
ENSDART00000065336
|
tmem14cb
|
transmembrane protein 14Cb |
| chr15_-_45446153 | 0.16 |
ENSDART00000100332
|
fgf12b
|
fibroblast growth factor 12b |
| chr7_-_28300008 | 0.16 |
|
|
|
| chr6_+_36861678 | 0.16 |
ENSDART00000104160
|
ENSDARG00000070746
|
ENSDARG00000070746 |
| chr6_-_54049582 | 0.16 |
|
|
|
| chr7_+_5788679 | 0.15 |
ENSDART00000173232
|
CU459186.7
|
ENSDARG00000105397 |
| chr3_-_32205873 | 0.15 |
ENSDART00000156918
|
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr14_+_47368951 | 0.15 |
ENSDART00000060577
|
tmem33
|
transmembrane protein 33 |
| chr6_+_40925259 | 0.15 |
ENSDART00000002728
ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr18_-_38263768 | 0.15 |
ENSDART00000076399
|
nat10
|
N-acetyltransferase 10 |
| chr5_-_31905091 | 0.14 |
ENSDART00000034705
|
ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr2_-_2252088 | 0.14 |
ENSDART00000164674
|
ss18
|
synovial sarcoma translocation, chromosome 18 (H. sapiens) |
| chr22_-_38947987 | 0.14 |
ENSDART00000008365
|
ncbp2
|
nuclear cap binding protein subunit 2 |
| chr23_-_24325912 | 0.14 |
|
|
|
| chr24_-_35819359 | 0.13 |
ENSDART00000165199
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr9_-_52951020 | 0.13 |
ENSDART00000169572
|
smarcal1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr15_-_8215831 | 0.13 |
ENSDART00000155381
|
bach1a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 a |
| chr22_+_38084309 | 0.13 |
ENSDART00000012212
|
commd2
|
COMM domain containing 2 |
| chr21_-_30123578 | 0.12 |
ENSDART00000126621
|
pfdn1
|
prefoldin subunit 1 |
| chr4_+_13587809 | 0.12 |
ENSDART00000138201
|
tnpo3
|
transportin 3 |
| chr20_-_55094208 | 0.12 |
ENSDART00000151522
|
si:dkey-15f23.1
|
si:dkey-15f23.1 |
| chr23_-_15234716 | 0.11 |
ENSDART00000158943
|
ndrg3b
|
ndrg family member 3b |
| chr7_-_57680717 | 0.11 |
|
|
|
| chr18_-_50108685 | 0.11 |
|
|
|
| chr12_-_11219985 | 0.11 |
ENSDART00000149229
|
si:ch73-30l9.1
|
si:ch73-30l9.1 |
| chr24_-_17261929 | 0.11 |
ENSDART00000153858
|
BX005392.2
|
ENSDARG00000096996 |
| chr20_+_31375056 | 0.11 |
|
|
|
| chr21_+_21158761 | 0.10 |
ENSDART00000058311
|
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr12_+_25132623 | 0.10 |
ENSDART00000127454
ENSDART00000122665 |
mta3
|
metastasis associated 1 family, member 3 |
| chr17_+_45892751 | 0.10 |
|
|
|
| chr5_+_41463969 | 0.10 |
ENSDART00000035235
|
UBB
|
si:ch211-202a12.4 |
| chr2_+_1825714 | 0.10 |
ENSDART00000148624
|
higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr5_-_22981395 | 0.10 |
ENSDART00000170293
|
si:dkeyp-20g2.1
|
si:dkeyp-20g2.1 |
| chr1_-_54366578 | 0.10 |
ENSDART00000170001
|
vps54
|
vacuolar protein sorting 54 homolog (S. cerevisiae) |
| chr17_-_15181427 | 0.09 |
ENSDART00000002176
ENSDART00000138764 |
wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr3_+_58175318 | 0.09 |
|
|
|
| chr11_-_2227358 | 0.09 |
ENSDART00000018131
|
hnrnpa1a
|
heterogeneous nuclear ribonucleoprotein A1a |
| chr21_+_43183835 | 0.09 |
ENSDART00000175107
|
aff4
|
AF4/FMR2 family, member 4 |
| chr2_-_57723042 | 0.09 |
ENSDART00000143973
|
sf3a2
|
splicing factor 3a, subunit 2 |
| chr17_+_25425967 | 0.09 |
ENSDART00000030691
|
clic4
|
chloride intracellular channel 4 |
| chr3_+_19536412 | 0.09 |
ENSDART00000006490
|
tlk2
|
tousled-like kinase 2 |
| chr1_-_48983245 | 0.09 |
|
|
|
| chr5_+_18421511 | 0.08 |
ENSDART00000051621
|
pgam5
|
PGAM family member 5, serine/threonine protein phosphatase, mitochondrial |
| chr2_-_24021423 | 0.08 |
ENSDART00000079131
|
cdyl
|
chromodomain protein, Y-like |
| chr23_-_29952057 | 0.08 |
ENSDART00000058407
|
slc25a33
|
solute carrier family 25 (pyrimidine nucleotide carrier), member 33 |
| chr11_+_15743714 | 0.08 |
ENSDART00000167191
|
pank4
|
pantothenate kinase 4 |
| chr15_-_33949567 | 0.08 |
ENSDART00000158445
|
pds5b
|
PDS5 cohesin associated factor B |
| chr15_-_34799968 | 0.07 |
ENSDART00000110964
|
bag6
|
BCL2-associated athanogene 6 |
| chr4_-_12478339 | 0.07 |
ENSDART00000027756
|
arhgef39
|
Rho guanine nucleotide exchange factor (GEF) 39 |
| chr4_-_62773293 | 0.07 |
ENSDART00000160787
|
znf1105
|
zinc finger protein 1105 |
| chr22_+_37696341 | 0.07 |
ENSDART00000007346
|
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr21_-_25235736 | 0.07 |
ENSDART00000158401
|
nfrkb
|
nuclear factor related to kappaB binding protein |
| chr11_-_22210506 | 0.07 |
ENSDART00000146873
|
tmem183a
|
transmembrane protein 183A |
| chr15_-_33949438 | 0.07 |
ENSDART00000158445
|
pds5b
|
PDS5 cohesin associated factor B |
| chr1_+_24960066 | 0.07 |
ENSDART00000054235
|
plrg1
|
pleiotropic regulator 1 |
| chr22_-_10026579 | 0.06 |
ENSDART00000179409
|
si:ch211-222k6.1
|
si:ch211-222k6.1 |
| chr20_+_31036284 | 0.06 |
ENSDART00000153344
|
sod2
|
superoxide dismutase 2, mitochondrial |
| chr7_+_23933847 | 0.06 |
ENSDART00000018580
|
nedd8
|
neural precursor cell expressed, developmentally down-regulated 8 |
| chr19_-_3947553 | 0.06 |
ENSDART00000172271
|
thrap3b
|
thyroid hormone receptor associated protein 3b |
| chr1_-_44970931 | 0.05 |
ENSDART00000042158
|
serhl
|
serine hydrolase-like |
| chr8_-_29921560 | 0.05 |
ENSDART00000125173
|
ercc6l2
|
excision repair cross-complementation group 6-like 2 |
| chr5_+_25133592 | 0.05 |
ENSDART00000098467
|
abhd17b
|
abhydrolase domain containing 17B |
| chr19_-_3947422 | 0.04 |
ENSDART00000172271
|
thrap3b
|
thyroid hormone receptor associated protein 3b |
| chr1_-_43300015 | 0.04 |
ENSDART00000138447
|
zgc:66472
|
zgc:66472 |
| chr6_+_34054187 | 0.04 |
ENSDART00000057732
|
ap1m3
|
adaptor-related protein complex 1, mu 3 subunit |
| chr6_+_19476528 | 0.04 |
ENSDART00000113911
|
micall1a
|
MICAL-like 1a |
| chr18_+_25491245 | 0.04 |
ENSDART00000174566
|
CR450716.2
|
ENSDARG00000108628 |
| chr5_-_20417926 | 0.04 |
ENSDART00000158030
|
si:ch211-225b11.4
|
si:ch211-225b11.4 |
| chr21_-_38106145 | 0.04 |
ENSDART00000151226
|
klf5l
|
Kruppel-like factor 5 like |
| chr3_+_27639256 | 0.03 |
ENSDART00000017962
|
eci1
|
enoyl-CoA delta isomerase 1 |
| chr8_+_28433869 | 0.03 |
ENSDART00000062716
|
slc52a3
|
solute carrier family 52 (riboflavin transporter), member 3 |
| chr19_+_46371722 | 0.03 |
ENSDART00000169710
|
utp23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr13_-_49826526 | 0.03 |
ENSDART00000099475
|
nid1a
|
nidogen 1a |
| chr2_+_31631321 | 0.02 |
ENSDART00000111038
|
ENSDARG00000077219
|
ENSDARG00000077219 |
| chr25_+_386868 | 0.02 |
|
|
|
| chr5_-_18420973 | 0.02 |
ENSDART00000041371
|
poli
|
polymerase (DNA directed) iota |
| chr25_+_28450465 | 0.02 |
ENSDART00000075151
|
amn1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
| chr20_+_23955892 | 0.02 |
ENSDART00000032393
|
ginm1
|
glycoprotein integral membrane 1 |
| chr17_+_1660922 | 0.02 |
ENSDART00000159190
|
srp14
|
signal recognition particle 14 |
| chr1_+_39848911 | 0.02 |
ENSDART00000137047
|
scoca
|
short coiled-coil protein a |
| chr14_+_25689596 | 0.02 |
ENSDART00000149087
|
slc36a1
|
solute carrier family 36 (proton/amino acid symporter), member 1 |
| chr9_-_33584043 | 0.02 |
ENSDART00000138844
|
caska
|
calcium/calmodulin-dependent serine protein kinase a |
| chr5_+_28170113 | 0.02 |
ENSDART00000114473
|
trafd1
|
TRAF-type zinc finger domain containing 1 |
| chr3_-_13311188 | 0.02 |
ENSDART00000080807
|
fbxw9
|
F-box and WD repeat domain containing 9 |
| chr19_+_2754903 | 0.01 |
ENSDART00000160533
|
tomm7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr20_+_7337738 | 0.01 |
ENSDART00000165596
|
dsg2.1
|
desmoglein 2, tandem duplicate 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0046833 | snRNA export from nucleus(GO:0006408) regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.1 | 0.4 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.1 | 0.9 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 1.4 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.3 | GO:0070655 | mechanosensory epithelium regeneration(GO:0070655) mechanoreceptor differentiation involved in mechanosensory epithelium regeneration(GO:0070656) neuromast regeneration(GO:0070657) neuromast hair cell differentiation involved in neuromast regeneration(GO:0070658) |
| 0.1 | 1.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.6 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 0.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 0.2 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.1 | 0.6 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.1 | 1.1 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.3 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.1 | 0.3 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 0.5 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.2 | GO:0000493 | box H/ACA snoRNP assembly(GO:0000493) |
| 0.1 | 0.2 | GO:0071846 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) actin filament debranching(GO:0071846) |
| 0.0 | 0.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.2 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.0 | 0.1 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.0 | 0.3 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.5 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.6 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.4 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.5 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.2 | GO:0050779 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.0 | 0.3 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 0.2 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 0.4 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.6 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.4 | GO:0034518 | RNA cap binding complex(GO:0034518) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.7 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.4 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.2 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 1.1 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.4 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 1.4 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 1.1 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 0.6 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 0.3 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.1 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.6 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.7 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.5 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.1 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.0 | 0.4 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.6 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.5 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.2 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |