Project
DANIO-CODE
Navigation
Downloads
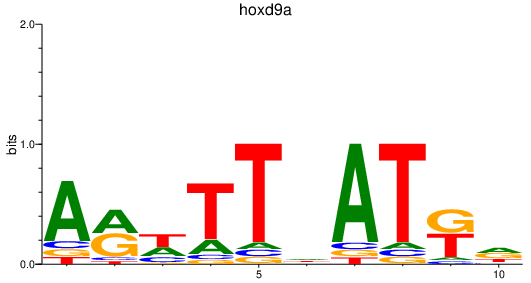
Results for hoxd9a
Z-value: 0.97
Transcription factors associated with hoxd9a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxd9a
|
ENSDARG00000059274 | homeobox D9a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxd9a | dr10_dc_chr9_-_1964814_1964941 | -0.17 | 5.2e-01 | Click! |
Activity profile of hoxd9a motif
Sorted Z-values of hoxd9a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxd9a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_28199458 | 2.52 |
ENSDART00000150330
|
buc
|
bucky ball |
| chr23_-_24416911 | 1.50 |
ENSDART00000166392
|
fam131c
|
family with sequence similarity 131, member C |
| chr15_-_33876041 | 1.25 |
ENSDART00000163249
|
zgc:73340
|
zgc:73340 |
| chr5_-_46028578 | 1.24 |
|
|
|
| chr2_+_11335733 | 1.19 |
ENSDART00000055737
|
tyw3
|
tRNA-yW synthesizing protein 3 homolog (S. cerevisiae) |
| chr14_-_15777250 | 1.08 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr24_+_16861121 | 1.00 |
ENSDART00000149149
|
zfx
|
zinc finger protein, X-linked |
| chr1_-_11191824 | 0.97 |
ENSDART00000163971
ENSDART00000123431 |
iqce
|
IQ motif containing E |
| chr16_+_35952050 | 0.97 |
ENSDART00000162411
|
sh3d21
|
SH3 domain containing 21 |
| chr24_-_32259029 | 0.91 |
ENSDART00000048633
|
trdmt1
|
tRNA aspartic acid methyltransferase 1 |
| chr22_-_3279707 | 0.85 |
ENSDART00000160305
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr16_-_17678748 | 0.85 |
ENSDART00000017142
|
m6pr
|
mannose-6-phosphate receptor (cation dependent) |
| chr13_+_33331767 | 0.83 |
ENSDART00000177841
|
zgc:136302
|
zgc:136302 |
| chr8_-_44617324 | 0.82 |
ENSDART00000063396
|
bag4
|
BCL2-associated athanogene 4 |
| chr5_+_3683209 | 0.82 |
ENSDART00000132056
|
ggnbp2
|
gametogenetin binding protein 2 |
| chr22_+_1837102 | 0.79 |
ENSDART00000163288
|
znf1174
|
zinc finger protein 1174 |
| chr15_+_22458649 | 0.77 |
ENSDART00000109931
|
oafa
|
OAF homolog a (Drosophila) |
| chr11_-_34521855 | 0.77 |
ENSDART00000039847
ENSDART00000135725 |
chchd4a
|
coiled-coil-helix-coiled-coil-helix domain containing 4a |
| chr13_-_28559160 | 0.73 |
ENSDART00000101582
|
pcgf6
|
polycomb group ring finger 6 |
| chr14_-_36004908 | 0.71 |
ENSDART00000077823
|
lrit3a
|
info leucine-rich repeat, immunoglobulin-like and transmembrane domains 3a |
| chr6_+_43501510 | 0.70 |
ENSDART00000154956
|
BX957311.1
|
ENSDARG00000097710 |
| chr11_-_30388949 | 0.69 |
ENSDART00000063775
|
ENSDARG00000043442
|
ENSDARG00000043442 |
| chr23_-_26241101 | 0.67 |
|
|
|
| chr4_+_377994 | 0.67 |
ENSDART00000148933
|
rpl18a
|
ribosomal protein L18a |
| chr23_+_16822435 | 0.66 |
ENSDART00000137737
|
fbxo44
|
F-box protein 44 |
| chr3_+_39437613 | 0.65 |
ENSDART00000055170
|
cln3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr2_-_6545431 | 0.64 |
ENSDART00000161934
|
si:dkey-119f1.1
|
si:dkey-119f1.1 |
| chr10_+_24530670 | 0.64 |
|
|
|
| chr4_+_11697088 | 0.63 |
ENSDART00000150669
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr14_-_33141111 | 0.63 |
ENSDART00000147059
|
lamp2
|
lysosomal-associated membrane protein 2 |
| chr5_+_31475897 | 0.62 |
ENSDART00000144510
|
zmat5
|
zinc finger, matrin-type 5 |
| chr2_-_11335644 | 0.62 |
ENSDART00000135450
|
cryz
|
crystallin, zeta (quinone reductase) |
| chr13_+_15669704 | 0.62 |
ENSDART00000146234
|
apopt1
|
apoptogenic 1, mitochondrial |
| chr19_-_4206333 | 0.62 |
ENSDART00000164611
|
rragcb
|
Ras-related GTP binding Cb |
| chr10_-_25448712 | 0.62 |
ENSDART00000140023
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr5_+_56896961 | 0.62 |
|
|
|
| chr2_+_30756137 | 0.61 |
ENSDART00000101861
|
tcea1
|
transcription elongation factor A (SII), 1 |
| chr12_+_18794467 | 0.59 |
ENSDART00000127536
|
cbx7b
|
chromobox homolog 7b |
| chr3_-_30985152 | 0.58 |
ENSDART00000058955
|
arl6ip1
|
ADP-ribosylation factor-like 6 interacting protein 1 |
| chr5_-_25791559 | 0.58 |
|
|
|
| chr12_+_19077980 | 0.58 |
ENSDART00000066393
|
pdap1a
|
pdgfa associated protein 1a |
| chr8_+_15213743 | 0.58 |
ENSDART00000063717
|
ENSDARG00000043402
|
ENSDARG00000043402 |
| chr20_-_22170411 | 0.58 |
ENSDART00000155568
|
BX088688.3
|
ENSDARG00000097598 |
| chr20_+_43794079 | 0.57 |
ENSDART00000045185
|
lin9
|
lin-9 DREAM MuvB core complex component |
| chr23_-_24416864 | 0.57 |
ENSDART00000166392
|
fam131c
|
family with sequence similarity 131, member C |
| chr22_+_35229352 | 0.57 |
ENSDART00000061315
ENSDART00000146430 |
tsc22d2
|
TSC22 domain family 2 |
| chr19_-_3833037 | 0.56 |
ENSDART00000168433
|
btr20
|
bloodthirsty-related gene family, member 20 |
| chr3_-_32741894 | 0.55 |
ENSDART00000075277
|
zgc:113090
|
zgc:113090 |
| chr15_-_25499679 | 0.55 |
ENSDART00000112079
|
tlcd2
|
TLC domain containing 2 |
| chr12_+_25341625 | 0.55 |
ENSDART00000011662
|
ppm1bb
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Bb |
| chr20_+_38555185 | 0.54 |
ENSDART00000020153
|
adck3
|
aarF domain containing kinase 3 |
| chr11_+_11319810 | 0.53 |
ENSDART00000162486
|
si:dkey-23f9.4
|
si:dkey-23f9.4 |
| chr4_-_20092548 | 0.53 |
ENSDART00000024647
|
dennd6b
|
DENN/MADD domain containing 6B |
| chr5_-_66687335 | 0.53 |
ENSDART00000062359
|
unga
|
uracil DNA glycosylase a |
| chr6_-_39922353 | 0.53 |
ENSDART00000065091
|
sumf1
|
sulfatase modifying factor 1 |
| chr18_+_44856321 | 0.52 |
ENSDART00000097328
|
arfgap2
|
ADP-ribosylation factor GTPase activating protein 2 |
| chr5_+_4906690 | 0.51 |
ENSDART00000124022
|
mapkap1
|
mitogen-activated protein kinase associated protein 1 |
| chr6_+_18941135 | 0.51 |
ENSDART00000044519
|
cx44.2
|
connexin 44.2 |
| chr11_+_30389309 | 0.51 |
ENSDART00000087909
|
tmem246
|
transmembrane protein 246 |
| chr1_-_11191863 | 0.50 |
ENSDART00000163971
ENSDART00000123431 |
iqce
|
IQ motif containing E |
| chr4_-_4603205 | 0.49 |
ENSDART00000130601
|
CABZ01020840.1
|
ENSDARG00000090401 |
| chr4_-_20092577 | 0.49 |
ENSDART00000164410
|
dennd6b
|
DENN/MADD domain containing 6B |
| chr13_+_15669924 | 0.49 |
ENSDART00000146234
|
apopt1
|
apoptogenic 1, mitochondrial |
| chr22_+_35229139 | 0.49 |
ENSDART00000061315
ENSDART00000146430 |
tsc22d2
|
TSC22 domain family 2 |
| chr9_+_2336680 | 0.49 |
ENSDART00000016417
|
atp5g3a
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9), genome duplicate a |
| chr19_+_7489293 | 0.49 |
ENSDART00000081746
|
apoa1bp
|
apolipoprotein A-I binding protein |
| chr19_-_431762 | 0.48 |
ENSDART00000165371
|
dus3l
|
dihydrouridine synthase 3-like (S. cerevisiae) |
| chr3_+_39437497 | 0.48 |
ENSDART00000055170
|
cln3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr11_-_19909409 | 0.48 |
ENSDART00000121722
|
si:dkey-274m17.3
|
si:dkey-274m17.3 |
| chr13_-_30741976 | 0.47 |
ENSDART00000142535
|
CR762493.1
|
ENSDARG00000092411 |
| chr4_-_71601845 | 0.46 |
ENSDART00000174158
|
CABZ01021433.1
|
ENSDARG00000105696 |
| chr5_-_52318158 | 0.46 |
ENSDART00000166267
|
zfpl1
|
zinc finger protein-like 1 |
| chr19_-_3832999 | 0.46 |
ENSDART00000168433
|
btr20
|
bloodthirsty-related gene family, member 20 |
| chr20_+_29663162 | 0.46 |
ENSDART00000062383
|
ywhaqa
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide a |
| chr15_-_23849212 | 0.45 |
ENSDART00000059354
|
rad1
|
RAD1 homolog (S. pombe) |
| chr14_+_7585289 | 0.45 |
ENSDART00000161307
|
ube2d2
|
ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) |
| chr1_-_343261 | 0.44 |
ENSDART00000010092
|
gas6
|
growth arrest-specific 6 |
| chr25_+_7067043 | 0.43 |
ENSDART00000179500
|
BX005076.1
|
ENSDARG00000107609 |
| chr20_-_29630048 | 0.43 |
ENSDART00000049224
|
taf1b
|
TATA box binding protein (Tbp)-associated factor, RNA polymerase I, B |
| chr12_-_33704272 | 0.43 |
ENSDART00000030566
|
galk1
|
galactokinase 1 |
| chr25_-_36512943 | 0.43 |
ENSDART00000114508
|
hprt1l
|
hypoxanthine phosphoribosyltransferase 1, like |
| chr13_-_24130160 | 0.42 |
ENSDART00000138747
ENSDART00000101168 |
urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr4_-_25192290 | 0.42 |
ENSDART00000066930
|
atp5c1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr8_+_44619220 | 0.42 |
ENSDART00000063392
|
lsm1
|
LSM1, U6 small nuclear RNA associated |
| chr12_+_19077847 | 0.42 |
ENSDART00000066393
|
pdap1a
|
pdgfa associated protein 1a |
| chr11_-_26463925 | 0.42 |
ENSDART00000083010
|
acad9
|
acyl-CoA dehydrogenase family, member 9 |
| chr16_+_27837465 | 0.42 |
|
|
|
| chr6_+_34406796 | 0.42 |
|
|
|
| chr2_+_35613093 | 0.42 |
ENSDART00000098734
|
cacybp
|
calcyclin binding protein |
| chr6_-_12434518 | 0.41 |
|
|
|
| chr1_+_22160932 | 0.41 |
ENSDART00000016488
|
gtf2e2
|
general transcription factor IIE, polypeptide 2, beta |
| chr19_-_2803212 | 0.41 |
ENSDART00000159253
|
exosc7
|
exosome component 7 |
| chr21_+_3013270 | 0.41 |
ENSDART00000160585
|
hnrnpabb
|
heterogeneous nuclear ribonucleoprotein A/Bb |
| chr3_-_39554099 | 0.40 |
ENSDART00000145303
|
b9d1
|
B9 protein domain 1 |
| chr20_+_36917979 | 0.40 |
ENSDART00000153085
|
heca
|
hdc homolog, cell cycle regulator |
| chr13_-_33348231 | 0.40 |
ENSDART00000160520
|
mad2l1bp
|
MAD2L1 binding protein |
| chr6_-_40654280 | 0.40 |
|
|
|
| chr19_+_40558066 | 0.40 |
ENSDART00000049968
|
si:ch211-173p18.3
|
si:ch211-173p18.3 |
| chr21_+_43177310 | 0.40 |
ENSDART00000121725
|
zcchc10
|
zinc finger, CCHC domain containing 10 |
| chr25_+_15176959 | 0.40 |
ENSDART00000045659
|
tcp11l1
|
t-complex 11, testis-specific-like 1 |
| chr21_+_39379031 | 0.40 |
ENSDART00000031470
|
pafah1b1b
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1b |
| chr18_+_47305642 | 0.39 |
ENSDART00000009775
|
rbm7
|
RNA binding motif protein 7 |
| chr9_-_32295031 | 0.39 |
ENSDART00000132448
|
BX530407.4
|
ENSDARG00000093950 |
| chr8_-_17755104 | 0.39 |
ENSDART00000131276
|
si:ch211-150o23.2
|
si:ch211-150o23.2 |
| chr12_+_9461872 | 0.38 |
ENSDART00000044150
|
dnajc9
|
DnaJ (Hsp40) homolog, subfamily C, member 9 |
| KN149787v1_-_20460 | 0.38 |
|
|
|
| chr5_-_3673698 | 0.38 |
|
|
|
| chr5_+_57151151 | 0.37 |
|
|
|
| chr15_-_26563295 | 0.37 |
|
|
|
| chr4_-_14955227 | 0.36 |
ENSDART00000038642
|
slc26a5
|
solute carrier family 26 (anion exchanger), member 5 |
| chr9_+_22970052 | 0.36 |
ENSDART00000143972
|
rif1
|
replication timing regulatory factor 1 |
| chr1_+_36593187 | 0.36 |
ENSDART00000171340
|
fam193a
|
family with sequence similarity 193, member A |
| chr14_+_47368995 | 0.36 |
ENSDART00000060577
|
tmem33
|
transmembrane protein 33 |
| chr14_-_35073975 | 0.36 |
ENSDART00000145033
|
rnaseh2c
|
ribonuclease H2, subunit C |
| chr25_+_7067115 | 0.36 |
ENSDART00000179500
|
BX005076.1
|
ENSDARG00000107609 |
| chr15_+_28242899 | 0.35 |
ENSDART00000037119
|
slc46a1
|
solute carrier family 46 (folate transporter), member 1 |
| chr12_-_20494169 | 0.35 |
ENSDART00000105362
|
snx11
|
sorting nexin 11 |
| chr17_-_51729568 | 0.35 |
ENSDART00000157244
|
exd2
|
exonuclease 3'-5' domain containing 2 |
| chr3_+_60299938 | 0.35 |
ENSDART00000053482
|
tsen54
|
TSEN54 tRNA splicing endonuclease subunit |
| chr9_-_12298632 | 0.35 |
ENSDART00000032344
|
nup35
|
nucleoporin 35 |
| chr10_+_10354740 | 0.34 |
ENSDART00000142895
|
urm1
|
ubiquitin related modifier 1 |
| KN150339v1_-_47655 | 0.34 |
|
|
|
| chr12_-_33704313 | 0.34 |
ENSDART00000030566
|
galk1
|
galactokinase 1 |
| chr16_+_13993746 | 0.34 |
ENSDART00000101304
|
atg12
|
ATG12 autophagy related 12 homolog (S. cerevisiae) |
| chr2_+_50071638 | 0.34 |
ENSDART00000056248
|
wdr48b
|
WD repeat domain 48b |
| chr4_+_70728870 | 0.34 |
ENSDART00000122809
|
si:ch211-161m3.2
|
si:ch211-161m3.2 |
| chr22_+_1643493 | 0.34 |
ENSDART00000167767
|
BX649416.1
|
ENSDARG00000098230 |
| chr4_+_5308883 | 0.34 |
ENSDART00000150366
|
si:ch211-214j24.10
|
si:ch211-214j24.10 |
| chr7_+_38306622 | 0.34 |
ENSDART00000074458
|
ptpmt1
|
protein tyrosine phosphatase, mitochondrial 1 |
| chr12_+_25341263 | 0.34 |
ENSDART00000153333
|
ppm1bb
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Bb |
| chr18_+_15137711 | 0.33 |
ENSDART00000128609
|
cry1ab
|
cryptochrome circadian clock 1ab |
| chr23_-_38228511 | 0.33 |
ENSDART00000087112
|
pfdn4
|
prefoldin subunit 4 |
| chr17_+_52526741 | 0.32 |
ENSDART00000109891
|
angel1
|
angel homolog 1 (Drosophila) |
| chr8_+_21101133 | 0.32 |
ENSDART00000056405
|
magoh
|
mago homolog, exon junction complex core component |
| chr3_+_41984535 | 0.32 |
ENSDART00000154052
|
ftsj2
|
FtsJ RNA methyltransferase homolog 2 (E. coli) |
| chr1_-_55195566 | 0.32 |
ENSDART00000019936
|
prkacab
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate b |
| chr15_+_17322173 | 0.31 |
ENSDART00000169608
|
dhx40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr5_+_68877574 | 0.31 |
ENSDART00000097244
|
gtf2h3
|
general transcription factor IIH, polypeptide 3 |
| chr7_-_6312243 | 0.31 |
ENSDART00000173045
|
si:ch1073-159d7.7
|
si:ch1073-159d7.7 |
| chr20_+_32598608 | 0.31 |
ENSDART00000021035
ENSDART00000152944 |
sec63
|
SEC63 homolog, protein translocation regulator |
| chr14_-_15776937 | 0.31 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr7_+_32626787 | 0.30 |
ENSDART00000140800
ENSDART00000075263 ENSDART00000137956 |
tssc4
|
tumor suppressing subtransferable candidate 4 |
| chr13_-_24129808 | 0.30 |
ENSDART00000138747
ENSDART00000101168 |
urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| KN149707v1_-_2099 | 0.30 |
|
|
|
| chr15_-_16936518 | 0.30 |
ENSDART00000142870
ENSDART00000171857 ENSDART00000152633 |
tyw1
|
tRNA-yW synthesizing protein 1 homolog (S. cerevisiae) |
| chr7_-_31523648 | 0.30 |
ENSDART00000169837
|
nap1l4b
|
nucleosome assembly protein 1-like 4b |
| chr5_-_41204170 | 0.30 |
ENSDART00000134492
|
alkbh2
|
alkB homolog 2, alpha-ketoglutarate-dependent dioxygenase |
| chr3_-_44414357 | 0.29 |
ENSDART00000160370
|
atp5h
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit d |
| chr4_-_20091845 | 0.29 |
|
|
|
| chr11_-_12744413 | 0.29 |
ENSDART00000104143
|
txlng
|
taxilin gamma |
| chr19_+_24484213 | 0.29 |
ENSDART00000100420
|
pex11b
|
peroxisomal biogenesis factor 11 beta |
| chr13_+_40628694 | 0.29 |
ENSDART00000143051
|
BX294129.1
|
ENSDARG00000092014 |
| chr3_-_30984999 | 0.29 |
ENSDART00000058955
|
arl6ip1
|
ADP-ribosylation factor-like 6 interacting protein 1 |
| chr22_+_12570345 | 0.29 |
ENSDART00000140054
|
ENSDARG00000041602
|
ENSDARG00000041602 |
| chr15_-_37687790 | 0.28 |
ENSDART00000154641
|
proser3
|
proline and serine rich 3 |
| chr14_+_47368951 | 0.28 |
ENSDART00000060577
|
tmem33
|
transmembrane protein 33 |
| chr8_-_26773755 | 0.28 |
ENSDART00000139787
|
kazna
|
kazrin, periplakin interacting protein a |
| chr21_+_39378835 | 0.28 |
ENSDART00000031470
|
pafah1b1b
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1b |
| chr20_-_54206786 | 0.28 |
ENSDART00000004756
|
hsp90aa1.1
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 1 |
| chr11_+_30005670 | 0.28 |
ENSDART00000167618
|
CR790368.1
|
ENSDARG00000100936 |
| chr10_+_25234636 | 0.27 |
ENSDART00000024514
|
chordc1b
|
cysteine and histidine-rich domain (CHORD) containing 1b |
| chr19_-_48327666 | 0.27 |
|
|
|
| chr7_+_23938868 | 0.27 |
ENSDART00000168257
|
CR318603.1
|
ENSDARG00000099215 |
| chr17_-_7194180 | 0.27 |
ENSDART00000098731
|
stxbp5b
|
syntaxin binding protein 5b (tomosyn) |
| chr9_-_25555518 | 0.27 |
ENSDART00000021672
|
epc2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr19_-_4206143 | 0.27 |
ENSDART00000164611
|
rragcb
|
Ras-related GTP binding Cb |
| chr10_-_32716462 | 0.27 |
ENSDART00000063544
|
atg101
|
autophagy related 101 |
| chr16_-_21816572 | 0.26 |
ENSDART00000115011
|
ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr22_-_22277386 | 0.26 |
ENSDART00000111711
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr12_+_22497249 | 0.26 |
ENSDART00000178120
|
CR848021.1
|
ENSDARG00000107088 |
| chr15_-_37687982 | 0.26 |
ENSDART00000154641
|
proser3
|
proline and serine rich 3 |
| chr10_+_11397605 | 0.26 |
ENSDART00000064215
|
cops4
|
COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis) |
| chr21_-_7322856 | 0.26 |
ENSDART00000151543
ENSDART00000114982 |
f2rl1.2
|
coagulation factor II (thrombin) receptor-like 1, tandem duplicate 2 |
| chr14_-_7585265 | 0.25 |
|
|
|
| chr5_-_15448501 | 0.25 |
ENSDART00000124467
|
xbp1
|
X-box binding protein 1 |
| chr4_+_5634076 | 0.25 |
ENSDART00000150847
|
mrps18a
|
mitochondrial ribosomal protein S18A |
| chr2_-_28199077 | 0.25 |
ENSDART00000121623
|
ENSDARG00000090770
|
ENSDARG00000090770 |
| chr3_+_32802124 | 0.25 |
ENSDART00000112742
|
nbr1b
|
neighbor of brca1 gene 1b |
| chr14_+_35074075 | 0.25 |
ENSDART00000084914
|
trmt12
|
tRNA methyltransferase 12 homolog (S. cerevisiae) |
| chr15_+_26667763 | 0.25 |
ENSDART00000155352
|
ENSDARG00000074561
|
ENSDARG00000074561 |
| chr24_-_6647047 | 0.24 |
|
|
|
| chr11_+_30389137 | 0.24 |
ENSDART00000087909
|
tmem246
|
transmembrane protein 246 |
| chr6_+_40925259 | 0.24 |
ENSDART00000002728
ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr15_+_37688139 | 0.24 |
ENSDART00000076066
|
lin37
|
lin-37 DREAM MuvB core complex component |
| chr4_-_15104842 | 0.24 |
ENSDART00000138183
|
nrf1
|
nuclear respiratory factor 1 |
| chr21_+_39918070 | 0.24 |
ENSDART00000135235
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr18_+_2902170 | 0.24 |
ENSDART00000170827
|
ccpg1
|
cell cycle progression 1 |
| chr5_+_56896748 | 0.24 |
|
|
|
| chr9_+_29806055 | 0.23 |
ENSDART00000143493
|
phf11
|
PHD finger protein 11 |
| chr3_-_26060787 | 0.23 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr15_-_37687938 | 0.22 |
ENSDART00000154641
|
proser3
|
proline and serine rich 3 |
| chr10_-_7862767 | 0.22 |
ENSDART00000059021
|
mat2aa
|
methionine adenosyltransferase II, alpha a |
| chr21_-_32748287 | 0.22 |
ENSDART00000031028
|
cnot6a
|
CCR4-NOT transcription complex, subunit 6a |
| chr21_+_39918125 | 0.22 |
ENSDART00000135235
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr5_-_23649439 | 0.22 |
|
|
|
| chr9_-_46614763 | 0.22 |
ENSDART00000009790
|
cx43.4
|
connexin 43.4 |
| chr3_-_39554155 | 0.21 |
ENSDART00000015393
|
b9d1
|
B9 protein domain 1 |
| chr9_+_33334773 | 0.21 |
ENSDART00000005879
|
atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr16_+_27449058 | 0.21 |
ENSDART00000132329
|
stx17
|
syntaxin 17 |
| chr4_+_5634025 | 0.21 |
ENSDART00000028941
|
mrps18a
|
mitochondrial ribosomal protein S18A |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.3 | 1.1 | GO:0015809 | arginine transport(GO:0015809) |
| 0.3 | 0.8 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 | 0.6 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.2 | 0.5 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.2 | 0.5 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.2 | 1.5 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.5 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.1 | 0.6 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.1 | 0.4 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.1 | 0.4 | GO:0006178 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine metabolic process(GO:0046098) guanine biosynthetic process(GO:0046099) hypoxanthine metabolic process(GO:0046100) |
| 0.1 | 0.4 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.1 | 0.3 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.1 | 0.4 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.3 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.7 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.1 | 0.3 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.1 | 0.4 | GO:0021693 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) |
| 0.1 | 0.3 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.1 | 0.3 | GO:0035511 | DNA dealkylation involved in DNA repair(GO:0006307) oxidative DNA demethylation(GO:0035511) |
| 0.1 | 0.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.3 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 0.3 | GO:0048785 | hatching gland development(GO:0048785) positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 0.9 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 0.8 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.1 | 1.2 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.7 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.3 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.4 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 1.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.3 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.3 | GO:0032447 | protein urmylation(GO:0032447) |
| 0.0 | 0.2 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.2 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.7 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.2 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.2 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 0.9 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 0.1 | GO:0072319 | clathrin coat disassembly(GO:0072318) vesicle uncoating(GO:0072319) |
| 0.0 | 0.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.4 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.6 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.1 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.8 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.4 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.2 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.3 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.3 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 0.6 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.3 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.0 | GO:0006683 | galactosylceramide catabolic process(GO:0006683) galactolipid catabolic process(GO:0019376) |
| 0.0 | 0.3 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.0 | 0.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.5 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.2 | 0.6 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 0.4 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 0.9 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.9 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.1 | 0.4 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.3 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) Sec62/Sec63 complex(GO:0031207) |
| 0.1 | 0.4 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.4 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 0.6 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 1.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.3 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.7 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.3 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.0 | 0.5 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.4 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.4 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.0 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.7 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.2 | 0.6 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 0.8 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.1 | 0.4 | GO:0052657 | guanine phosphoribosyltransferase activity(GO:0052657) |
| 0.1 | 0.5 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.1 | 0.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.3 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.1 | 0.5 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.5 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.1 | 0.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.4 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.1 | 0.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.7 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.6 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.4 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 1.3 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.5 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.3 | GO:0004691 | AMP-activated protein kinase activity(GO:0004679) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.3 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.6 | GO:0008026 | ATP-dependent DNA helicase activity(GO:0004003) ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 0.2 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 1.4 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.2 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.0 | 0.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.9 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.1 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.0 | 0.3 | GO:0034595 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.0 | 0.2 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 1.4 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.6 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.3 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) macromolecule transmembrane transporter activity(GO:0022884) |
| 0.0 | 0.7 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.0 | GO:0004336 | galactosylceramidase activity(GO:0004336) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.8 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.5 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.2 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 0.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.8 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.5 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.4 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.2 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.8 | REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |
| 0.0 | 0.4 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.5 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |