Project
DANIO-CODE
Navigation
Downloads
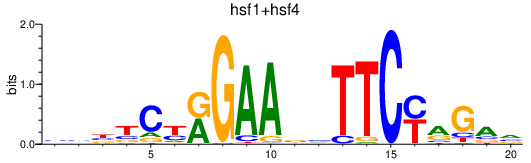
Results for hsf1+hsf4
Z-value: 0.67
Transcription factors associated with hsf1+hsf4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hsf1
|
ENSDARG00000008818 | heat shock transcription factor 1 |
|
hsf4
|
ENSDARG00000013251 | heat shock transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hsf4 | dr10_dc_chr18_-_22764560_22764583 | -0.74 | 1.1e-03 | Click! |
| hsf1 | dr10_dc_chr19_+_3115685_3115724 | 0.60 | 1.5e-02 | Click! |
Activity profile of hsf1+hsf4 motif
Sorted Z-values of hsf1+hsf4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hsf1+hsf4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_43749940 | 2.17 |
ENSDART00000133317
ENSDART00000140316 ENSDART00000142929 ENSDART00000133874 |
zgc:66313
|
zgc:66313 |
| chr1_-_21961282 | 2.13 |
ENSDART00000171830
|
uchl1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr17_-_2419079 | 1.75 |
ENSDART00000065821
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr13_-_28480530 | 1.64 |
ENSDART00000043156
|
cyp17a1
|
cytochrome P450, family 17, subfamily A, polypeptide 1 |
| chr9_+_8418408 | 1.53 |
ENSDART00000043067
|
zgc:171776
|
zgc:171776 |
| chr22_+_4145005 | 1.49 |
ENSDART00000171774
|
zgc:171566
|
zgc:171566 |
| chr10_+_15066791 | 1.49 |
ENSDART00000140084
|
si:dkey-88l16.5
|
si:dkey-88l16.5 |
| chr5_-_66145078 | 1.39 |
ENSDART00000041441
|
stip1
|
stress-induced phosphoprotein 1 |
| chr9_+_28293831 | 1.31 |
ENSDART00000101338
|
idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr3_+_36093650 | 1.30 |
ENSDART00000150994
|
dus1l
|
dihydrouridine synthase 1-like (S. cerevisiae) |
| chr3_+_13450981 | 1.24 |
ENSDART00000162124
|
hspbp1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr19_+_14295978 | 1.23 |
ENSDART00000168260
|
nudc
|
nudC nuclear distribution protein |
| chr2_-_57339717 | 1.23 |
ENSDART00000150034
|
pias4b
|
protein inhibitor of activated STAT, 4b |
| chr3_-_10044965 | 1.18 |
ENSDART00000154460
|
FP243382.1
|
ENSDARG00000097649 |
| chr22_+_2014357 | 1.17 |
ENSDART00000165168
|
znf1157
|
zinc finger protein 1157 |
| chr22_+_2014149 | 1.15 |
ENSDART00000165168
|
znf1157
|
zinc finger protein 1157 |
| chr15_+_44168257 | 1.12 |
ENSDART00000176254
|
CU655961.7
|
ENSDARG00000106724 |
| chr3_-_10045028 | 1.10 |
ENSDART00000154460
|
FP243382.1
|
ENSDARG00000097649 |
| chr21_+_13291305 | 1.10 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr18_+_43039021 | 1.07 |
|
|
|
| chr17_+_52526741 | 1.05 |
ENSDART00000109891
|
angel1
|
angel homolog 1 (Drosophila) |
| chr13_-_28480153 | 0.98 |
ENSDART00000043156
|
cyp17a1
|
cytochrome P450, family 17, subfamily A, polypeptide 1 |
| chr21_+_13291242 | 0.93 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr21_+_38685219 | 0.83 |
ENSDART00000076157
|
rab24
|
RAB24, member RAS oncogene family |
| chr7_-_2614300 | 0.82 |
ENSDART00000153548
|
BX323987.1
|
ENSDARG00000097745 |
| chr2_-_21967353 | 0.82 |
ENSDART00000137169
|
zgc:55781
|
zgc:55781 |
| chr2_+_19514886 | 0.81 |
ENSDART00000166073
|
elovl1a
|
ELOVL fatty acid elongase 1a |
| chr17_-_43749763 | 0.81 |
ENSDART00000167422
|
ahsa1b
|
AHA1, activator of heat shock protein ATPase homolog 1b |
| chr21_-_2261849 | 0.78 |
ENSDART00000172041
ENSDART00000161554 |
si:ch73-299h12.4
|
si:ch73-299h12.4 |
| chr15_-_30247084 | 0.77 |
ENSDART00000154069
|
BX322555.4
|
ENSDARG00000097908 |
| chr19_+_14295836 | 0.77 |
ENSDART00000170335
|
nudc
|
nudC nuclear distribution protein |
| chr15_+_44909293 | 0.74 |
ENSDART00000111373
|
pdgfd
|
platelet derived growth factor d |
| chr23_+_2481946 | 0.72 |
ENSDART00000126038
|
tcp1
|
t-complex 1 |
| chr6_-_52725402 | 0.71 |
ENSDART00000033949
|
oser1
|
oxidative stress responsive serine-rich 1 |
| chr10_-_29945491 | 0.71 |
ENSDART00000063923
ENSDART00000136264 |
zpr1
|
ZPR1 zinc finger |
| chr13_-_44882023 | 0.71 |
|
|
|
| chr13_-_28480365 | 0.70 |
ENSDART00000043156
|
cyp17a1
|
cytochrome P450, family 17, subfamily A, polypeptide 1 |
| chr10_+_15066654 | 0.67 |
ENSDART00000135667
|
si:dkey-88l16.5
|
si:dkey-88l16.5 |
| chr21_+_13291347 | 0.65 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr22_-_31089831 | 0.65 |
|
|
|
| chr22_+_2014232 | 0.65 |
ENSDART00000165168
|
znf1157
|
zinc finger protein 1157 |
| chr17_+_14956899 | 0.64 |
|
|
|
| chr14_-_6637670 | 0.64 |
ENSDART00000126279
|
clk4a
|
CDC-like kinase 4a |
| chr16_+_37716167 | 0.63 |
|
|
|
| chr20_-_35601497 | 0.63 |
|
|
|
| chr3_+_13450861 | 0.63 |
ENSDART00000162124
|
hspbp1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr15_-_19396871 | 0.62 |
|
|
|
| chr24_+_36430199 | 0.62 |
|
|
|
| chr12_-_10384031 | 0.61 |
ENSDART00000052004
|
zgc:153595
|
zgc:153595 |
| chr7_-_40359613 | 0.55 |
ENSDART00000173670
|
ube3c
|
ubiquitin protein ligase E3C |
| chr22_+_20686657 | 0.55 |
|
|
|
| chr6_-_39633603 | 0.54 |
ENSDART00000104042
|
atf7b
|
activating transcription factor 7b |
| chr17_-_23201467 | 0.54 |
ENSDART00000156411
|
fam98a
|
family with sequence similarity 98, member A |
| chr17_+_19711077 | 0.53 |
|
|
|
| chr12_-_10383960 | 0.52 |
ENSDART00000052004
|
zgc:153595
|
zgc:153595 |
| chr12_-_18839743 | 0.52 |
ENSDART00000172574
|
ep300a
|
E1A binding protein p300 a |
| chr11_-_11641577 | 0.52 |
ENSDART00000161821
|
si:dkey-28e7.3
|
si:dkey-28e7.3 |
| chr16_+_37716207 | 0.51 |
|
|
|
| chr17_+_19711104 | 0.50 |
|
|
|
| chr9_+_8418549 | 0.50 |
ENSDART00000043067
|
zgc:171776
|
zgc:171776 |
| chr7_+_30011202 | 0.49 |
ENSDART00000108782
|
polr2m
|
RNA polymerase II subunit M |
| chr6_-_29169308 | 0.49 |
ENSDART00000110288
ENSDART00000008841 |
zbtb11
|
zinc finger and BTB domain containing 11 |
| chr15_+_44909356 | 0.49 |
ENSDART00000111373
|
pdgfd
|
platelet derived growth factor d |
| chr17_-_29005489 | 0.48 |
ENSDART00000157210
|
CR925802.1
|
ENSDARG00000097263 |
| chr21_-_2261748 | 0.48 |
ENSDART00000172041
ENSDART00000161554 |
si:ch73-299h12.4
|
si:ch73-299h12.4 |
| chr5_-_3255080 | 0.47 |
ENSDART00000152108
|
hspb1
|
heat shock protein, alpha-crystallin-related, 1 |
| chr9_-_7676688 | 0.46 |
ENSDART00000102706
|
dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr8_+_9955720 | 0.45 |
|
|
|
| chr14_+_533611 | 0.45 |
|
|
|
| chr3_-_32158331 | 0.44 |
ENSDART00000055295
ENSDART00000156243 |
cpt1cb
|
carnitine palmitoyltransferase 1Cb |
| chr22_-_20925422 | 0.43 |
ENSDART00000002029
|
fkbp8
|
FK506 binding protein 8 |
| chr10_+_29963102 | 0.43 |
ENSDART00000168898
|
hspa8
|
heat shock protein 8 |
| chr8_+_13757024 | 0.42 |
|
|
|
| chr15_+_44168674 | 0.42 |
|
|
|
| chr24_+_32610430 | 0.42 |
ENSDART00000132417
|
yme1l1a
|
YME1-like 1a |
| chr14_-_36004835 | 0.42 |
ENSDART00000077823
|
lrit3a
|
info leucine-rich repeat, immunoglobulin-like and transmembrane domains 3a |
| chr15_-_46864317 | 0.41 |
|
|
|
| chr11_-_18439280 | 0.39 |
|
|
|
| chr10_+_29964045 | 0.39 |
ENSDART00000118838
|
SNORD14
|
Small nucleolar RNA SNORD14 |
| chr8_+_13757180 | 0.39 |
|
|
|
| chr3_-_43134137 | 0.38 |
ENSDART00000165592
|
rmi2
|
RecQ mediated genome instability 2 |
| chr2_-_56632969 | 0.37 |
ENSDART00000089158
|
hmha1a
|
histocompatibility (minor) HA-1 a |
| chr16_-_22397159 | 0.37 |
|
|
|
| chr7_+_73583860 | 0.36 |
ENSDART00000166244
|
si:ch73-252p3.1
|
si:ch73-252p3.1 |
| chr4_+_74873994 | 0.35 |
ENSDART00000160218
ENSDART00000110144 |
zgc:113119
|
zgc:113119 |
| chr18_+_37279099 | 0.35 |
ENSDART00000135444
|
tbcb
|
tubulin folding cofactor B |
| chr17_+_19711153 | 0.34 |
|
|
|
| chr2_+_10209233 | 0.34 |
ENSDART00000160304
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr19_+_46667326 | 0.34 |
ENSDART00000158032
|
grinaa
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1a (glutamate binding) |
| chr24_+_32610480 | 0.33 |
ENSDART00000132417
|
yme1l1a
|
YME1-like 1a |
| chr24_+_32635139 | 0.32 |
ENSDART00000175986
|
CU138525.1
|
ENSDARG00000105889 |
| chr14_+_35552581 | 0.31 |
ENSDART00000135079
|
zgc:63568
|
zgc:63568 |
| chr15_-_34732837 | 0.30 |
ENSDART00000003674
|
ankmy2a
|
ankyrin repeat and MYND domain containing 2a |
| chr10_+_44846631 | 0.30 |
|
|
|
| chr8_-_6782167 | 0.28 |
ENSDART00000135834
ENSDART00000172157 |
dock5
|
dedicator of cytokinesis 5 |
| chr5_-_3254995 | 0.28 |
ENSDART00000152108
|
hspb1
|
heat shock protein, alpha-crystallin-related, 1 |
| chr17_+_4903106 | 0.28 |
ENSDART00000064313
ENSDART00000121806 |
cdc5l
|
CDC5 cell division cycle 5-like (S. pombe) |
| chr18_+_37279031 | 0.27 |
ENSDART00000135444
|
tbcb
|
tubulin folding cofactor B |
| chr22_-_36705684 | 0.27 |
|
|
|
| chr5_+_32698516 | 0.26 |
ENSDART00000097945
|
usp20
|
ubiquitin specific peptidase 20 |
| chr5_-_13278406 | 0.25 |
ENSDART00000051655
|
snrnp27
|
small nuclear ribonucleoprotein 27 (U4/U6.U5) |
| chr23_+_12427170 | 0.25 |
ENSDART00000143728
|
pigt
|
phosphatidylinositol glycan anchor biosynthesis, class T |
| chr5_+_41463969 | 0.24 |
ENSDART00000035235
|
UBB
|
si:ch211-202a12.4 |
| chr1_-_53081805 | 0.23 |
ENSDART00000026409
|
cct4
|
chaperonin containing TCP1, subunit 4 (delta) |
| chr9_+_38233505 | 0.23 |
ENSDART00000022574
|
stam2
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 |
| chr2_-_56632913 | 0.21 |
ENSDART00000089158
|
hmha1a
|
histocompatibility (minor) HA-1 a |
| chr3_-_43134049 | 0.20 |
ENSDART00000165592
|
rmi2
|
RecQ mediated genome instability 2 |
| chr8_-_985916 | 0.18 |
ENSDART00000170737
|
smyd1b
|
SET and MYND domain containing 1b |
| chr22_-_37630428 | 0.18 |
ENSDART00000149482
|
fxr1
|
fragile X mental retardation, autosomal homolog 1 |
| chr22_+_34726990 | 0.17 |
ENSDART00000082066
|
atpv0e2
|
ATPase, H+ transporting V0 subunit e2 |
| chr10_+_29963998 | 0.17 |
ENSDART00000118838
|
SNORD14
|
Small nucleolar RNA SNORD14 |
| chr21_-_2107554 | 0.16 |
ENSDART00000159378
|
AL627305.2
|
ENSDARG00000101832 |
| chr18_+_37278733 | 0.16 |
ENSDART00000135444
|
tbcb
|
tubulin folding cofactor B |
| chr6_+_55835894 | 0.16 |
ENSDART00000108786
|
si:ch211-81n22.1
|
si:ch211-81n22.1 |
| chr12_-_17029512 | 0.14 |
|
|
|
| chr2_+_42286232 | 0.14 |
ENSDART00000008268
|
cyp7b1
|
cytochrome P450, family 7, subfamily B, polypeptide 1 |
| chr23_+_27957315 | 0.14 |
|
|
|
| chr17_+_53209709 | 0.14 |
ENSDART00000158313
|
ddx24
|
DEAD (Asp-Glu-Ala-Asp) box helicase 24 |
| chr1_+_43352429 | 0.13 |
ENSDART00000124873
|
si:ch73-109d9.4
|
si:ch73-109d9.4 |
| chr18_+_27355569 | 0.11 |
ENSDART00000136172
|
si:dkey-29p10.4
|
si:dkey-29p10.4 |
| chr2_-_39049391 | 0.10 |
ENSDART00000044331
|
copb2
|
coatomer protein complex, subunit beta 2 |
| chr9_-_30752583 | 0.09 |
|
|
|
| chr21_+_43490314 | 0.08 |
ENSDART00000026666
|
atg4a
|
autophagy related 4A, cysteine peptidase |
| chr20_-_33802212 | 0.07 |
ENSDART00000166573
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr6_+_7093685 | 0.07 |
ENSDART00000177226
|
dzip1
|
DAZ interacting zinc finger protein 1 |
| chr10_+_29964164 | 0.06 |
ENSDART00000118838
|
SNORD14
|
Small nucleolar RNA SNORD14 |
| chr5_-_13278451 | 0.05 |
ENSDART00000051655
|
snrnp27
|
small nuclear ribonucleoprotein 27 (U4/U6.U5) |
| chr6_-_13881001 | 0.04 |
ENSDART00000004656
|
ENSDARG00000020187
|
ENSDARG00000020187 |
| chr13_-_50032548 | 0.03 |
ENSDART00000038120
|
cacul1
|
CDK2 associated cullin domain 1 |
| chr14_-_220808 | 0.03 |
|
|
|
| chr24_-_39970897 | 0.02 |
ENSDART00000134251
|
slc12a7b
|
solute carrier family 12 (potassium/chloride transporter), member 7b |
| chr22_-_8276968 | 0.01 |
ENSDART00000123982
|
CABZ01077218.1
|
ENSDARG00000089720 |
| chr25_-_252714 | 0.01 |
|
|
|
| chr3_-_32409303 | 0.01 |
ENSDART00000151476
|
rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr9_-_21427035 | 0.01 |
ENSDART00000146764
|
cox17
|
COX17 cytochrome c oxidase copper chaperone |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:1903449 | male sex determination(GO:0030238) androst-4-ene-3,17-dione biosynthetic process(GO:1903449) |
| 0.4 | 1.3 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.2 | 1.5 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.2 | 1.2 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 0.7 | GO:0046685 | regulation of myofibril size(GO:0014881) response to arsenic-containing substance(GO:0046685) |
| 0.1 | 0.7 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.1 | 0.5 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.1 | 0.6 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 1.8 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.4 | GO:1902946 | protein localization to early endosome(GO:1902946) |
| 0.1 | 0.8 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.1 | 1.2 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.7 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.8 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.3 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.0 | 0.3 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.3 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.5 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 1.6 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.2 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 1.3 | GO:0008033 | tRNA processing(GO:0008033) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 0.2 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.3 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.8 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.3 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.4 | 1.3 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.3 | 1.3 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.2 | 1.8 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.2 | 1.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.2 | 1.5 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.2 | 1.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 2.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 1.9 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.7 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.8 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 1.0 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 1.8 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 2.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 2.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 0.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 1.2 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.4 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 1.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 0.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 2.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.4 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 1.2 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.7 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |