Project
DANIO-CODE
Navigation
Downloads
Results for hsf2+si:dkey-18a10.3
Z-value: 1.72
Transcription factors associated with hsf2+si:dkey-18a10.3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hsf2
|
ENSDARG00000053097 | heat shock transcription factor 2 |
|
si_dkey-18a10.3
|
ENSDARG00000090814 | si_dkey-18a10.3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hsf2 | dr10_dc_chr20_-_40589999_40590133 | -0.65 | 6.0e-03 | Click! |
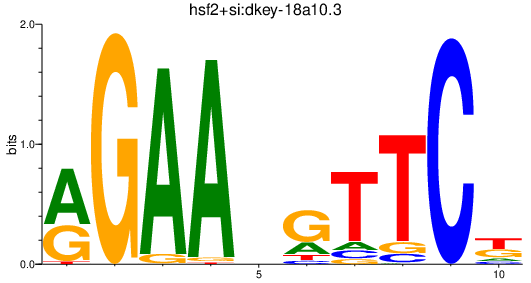
Activity profile of hsf2+si:dkey-18a10.3 motif
Sorted Z-values of hsf2+si:dkey-18a10.3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hsf2+si:dkey-18a10.3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_54206610 | 8.82 |
ENSDART00000004756
|
hsp90aa1.1
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 1 |
| chr15_+_37039861 | 7.17 |
ENSDART00000172664
|
kirrel3l
|
kin of IRRE like 3 like |
| chr23_-_31446156 | 6.05 |
ENSDART00000053367
|
hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr9_-_11589126 | 4.86 |
ENSDART00000146832
|
cryba2b
|
crystallin, beta A2b |
| chr2_+_36634309 | 4.54 |
ENSDART00000169547
ENSDART00000098417 |
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr20_-_44598129 | 4.47 |
ENSDART00000012229
|
fkbp1b
|
FK506 binding protein 1b |
| chr7_-_50827308 | 4.26 |
ENSDART00000121574
|
col4a6
|
collagen, type IV, alpha 6 |
| chr22_-_3661536 | 4.09 |
ENSDART00000153634
|
FP565432.1
|
ENSDARG00000097145 |
| chr12_+_15235008 | 4.04 |
|
|
|
| chr7_+_38479571 | 3.79 |
ENSDART00000170486
|
f2
|
coagulation factor II (thrombin) |
| chr3_-_23444371 | 3.76 |
ENSDART00000087726
|
igf2bp1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr20_+_15653121 | 3.76 |
ENSDART00000152702
|
jun
|
jun proto-oncogene |
| chr6_+_49096966 | 3.65 |
ENSDART00000141042
ENSDART00000143369 |
slc25a22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr11_-_41219340 | 3.45 |
ENSDART00000109204
|
megf6b
|
multiple EGF-like-domains 6b |
| chr4_-_17066886 | 3.43 |
ENSDART00000134595
|
sox5
|
SRY (sex determining region Y)-box 5 |
| chr6_+_16279737 | 3.40 |
ENSDART00000040035
|
ccdc80l1
|
coiled-coil domain containing 80 like 1 |
| chr25_+_33833810 | 3.30 |
ENSDART00000073440
|
ENSDARG00000051762
|
ENSDARG00000051762 |
| chr8_-_1904720 | 3.24 |
ENSDART00000081563
|
si:dkey-178e17.3
|
si:dkey-178e17.3 |
| chr13_+_24703802 | 3.19 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr23_+_36079164 | 3.13 |
ENSDART00000103131
|
hoxc1a
|
homeo box C1a |
| chr22_+_24362712 | 2.87 |
ENSDART00000157861
|
p3h2
|
prolyl 3-hydroxylase 2 |
| chr2_-_58111727 | 2.85 |
ENSDART00000004431
ENSDART00000166845 ENSDART00000163999 |
epb41l3b
|
erythrocyte membrane protein band 4.1-like 3b |
| chr7_-_60525749 | 2.66 |
ENSDART00000136999
|
pcxb
|
pyruvate carboxylase b |
| chr23_+_36079624 | 2.65 |
ENSDART00000103131
|
hoxc1a
|
homeo box C1a |
| chr10_+_35478939 | 2.63 |
ENSDART00000147303
|
hhla2a.1
|
HERV-H LTR-associating 2a, tandem duplicate 1 |
| chr22_+_21997000 | 2.60 |
ENSDART00000046174
|
gna15.1
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 1 |
| chr19_-_18316039 | 2.59 |
ENSDART00000151133
|
top2b
|
topoisomerase (DNA) II beta |
| chr14_+_27936303 | 2.50 |
ENSDART00000003293
|
mid2
|
midline 2 |
| chr23_-_24755654 | 2.39 |
ENSDART00000104035
|
ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr2_-_35584440 | 2.35 |
ENSDART00000125298
|
tnw
|
tenascin W |
| chr1_+_1953714 | 2.33 |
ENSDART00000164488
ENSDART00000167050 ENSDART00000122626 ENSDART00000128187 |
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr16_-_8481813 | 2.31 |
ENSDART00000051166
|
grb10b
|
growth factor receptor-bound protein 10b |
| chr6_-_30876091 | 2.28 |
ENSDART00000155330
|
pde4ba
|
phosphodiesterase 4B, cAMP-specific a |
| chr16_+_34569479 | 2.26 |
ENSDART00000041007
|
stmn1b
|
stathmin 1b |
| chr16_-_44575942 | 2.20 |
|
|
|
| chr22_-_14090538 | 2.15 |
ENSDART00000105717
|
aox5
|
aldehyde oxidase 5 |
| chr1_+_9999842 | 2.10 |
ENSDART00000152438
|
ENSDARG00000037679
|
ENSDARG00000037679 |
| chr16_-_17164459 | 2.05 |
ENSDART00000178443
|
tnfrsf1a
|
tumor necrosis factor receptor superfamily, member 1a |
| chr8_+_584353 | 2.04 |
ENSDART00000048498
|
CABZ01049925.1
|
ENSDARG00000079790 |
| chr10_-_10906027 | 2.03 |
ENSDART00000122657
|
nrarpa
|
notch-regulated ankyrin repeat protein a |
| chr13_-_45338347 | 2.03 |
ENSDART00000043345
|
rsrp1
|
arginine/serine-rich protein 1 |
| chr4_-_13394059 | 1.99 |
ENSDART00000175909
ENSDART00000174861 |
BX511038.1
|
ENSDARG00000108089 |
| chr6_-_39311711 | 1.95 |
|
|
|
| chr9_-_22507592 | 1.90 |
ENSDART00000132029
|
crygm2d2
|
crystallin, gamma M2d2 |
| chr5_+_32191063 | 1.88 |
ENSDART00000077189
|
ier5l
|
immediate early response 5-like |
| chr20_+_23599157 | 1.86 |
ENSDART00000149922
|
palld
|
palladin, cytoskeletal associated protein |
| chr16_+_5466342 | 1.84 |
ENSDART00000160008
|
plecb
|
plectin b |
| chr20_+_34640435 | 1.83 |
ENSDART00000152073
|
ENSDARG00000054723
|
ENSDARG00000054723 |
| chr5_-_20375703 | 1.73 |
ENSDART00000134697
|
pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr25_+_27829693 | 1.73 |
ENSDART00000010325
|
fezf1
|
FEZ family zinc finger 1 |
| chr1_+_45961348 | 1.70 |
ENSDART00000053221
|
arl11
|
ADP-ribosylation factor-like 11 |
| chr15_+_37040064 | 1.66 |
ENSDART00000172664
|
kirrel3l
|
kin of IRRE like 3 like |
| chr10_+_9575 | 1.63 |
|
|
|
| chr17_+_15026344 | 1.63 |
ENSDART00000154987
|
samd4a
|
sterile alpha motif domain containing 4A |
| chr4_-_24298688 | 1.60 |
ENSDART00000077926
ENSDART00000128368 |
celf2
|
cugbp, Elav-like family member 2 |
| chr22_+_24362832 | 1.59 |
ENSDART00000157861
|
p3h2
|
prolyl 3-hydroxylase 2 |
| chr15_-_37949371 | 1.56 |
ENSDART00000031418
|
hsc70
|
heat shock cognate 70 |
| chr4_-_72603830 | 1.56 |
ENSDART00000168547
|
si:cabz01071912.2
|
si:cabz01071912.2 |
| chr3_+_33213672 | 1.56 |
ENSDART00000162870
|
mpp2a
|
membrane protein, palmitoylated 2a (MAGUK p55 subfamily member 2) |
| chr9_+_32490721 | 1.53 |
ENSDART00000078608
|
hspe1
|
heat shock 10 protein 1 |
| chr19_-_36009321 | 1.51 |
ENSDART00000136811
|
col8a2
|
collagen, type VIII, alpha 2 |
| chr6_+_13099562 | 1.49 |
ENSDART00000038505
|
rprmb
|
reprimo, TP53 dependent G2 arrest mediator candidate b |
| chr6_-_60152594 | 1.47 |
ENSDART00000083453
|
slc32a1
|
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr5_-_13184573 | 1.46 |
ENSDART00000018351
|
zgc:65851
|
zgc:65851 |
| chr18_+_14725148 | 1.46 |
ENSDART00000146128
|
uri1
|
URI1, prefoldin-like chaperone |
| chr19_+_20194594 | 1.43 |
ENSDART00000169074
|
hoxa4a
|
homeobox A4a |
| chr11_+_29753417 | 1.42 |
|
|
|
| chr5_+_41492032 | 1.39 |
ENSDART00000171678
|
ubb
|
ubiquitin B |
| chr24_-_39970858 | 1.37 |
ENSDART00000134251
|
slc12a7b
|
solute carrier family 12 (potassium/chloride transporter), member 7b |
| chr2_+_52561735 | 1.37 |
ENSDART00000175043
|
CABZ01040021.1
|
ENSDARG00000107973 |
| chr10_+_29966381 | 1.36 |
ENSDART00000141549
|
hspa8
|
heat shock protein 8 |
| chr23_+_45876483 | 1.36 |
ENSDART00000169521
ENSDART00000162915 |
dclk2b
|
doublecortin-like kinase 2b |
| chr17_+_41475052 | 1.33 |
ENSDART00000075331
|
insm1b
|
insulinoma-associated 1b |
| chr11_-_41219410 | 1.32 |
ENSDART00000109204
|
megf6b
|
multiple EGF-like-domains 6b |
| chr20_-_54206786 | 1.32 |
ENSDART00000004756
|
hsp90aa1.1
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 1 |
| chr21_-_91068 | 1.30 |
|
|
|
| chr3_-_53467621 | 1.26 |
ENSDART00000032788
|
rdh8a
|
retinol dehydrogenase 8a |
| chr6_+_43402753 | 1.25 |
ENSDART00000143374
|
mitfa
|
microphthalmia-associated transcription factor a |
| chr4_+_21091640 | 1.22 |
ENSDART00000005847
|
nav3
|
neuron navigator 3 |
| chr13_+_25356184 | 1.21 |
ENSDART00000057689
|
bag3
|
BCL2-associated athanogene 3 |
| chr13_+_25356121 | 1.20 |
ENSDART00000057689
|
bag3
|
BCL2-associated athanogene 3 |
| chr2_+_36634251 | 1.16 |
ENSDART00000169547
ENSDART00000098417 |
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr25_+_34745872 | 1.15 |
ENSDART00000003494
|
slc17a6a
|
solute carrier family 17 (vesicular glutamate transporter), member 6a |
| chr2_+_31974269 | 1.14 |
ENSDART00000012413
|
ankhb
|
ANKH inorganic pyrophosphate transport regulator b |
| chr6_-_60152693 | 1.14 |
ENSDART00000083453
|
slc32a1
|
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr5_+_19599323 | 1.12 |
ENSDART00000079424
|
si:rp71-1c23.3
|
si:rp71-1c23.3 |
| chr10_+_29966252 | 1.11 |
ENSDART00000141549
|
hspa8
|
heat shock protein 8 |
| chr3_-_5318206 | 1.10 |
ENSDART00000137105
|
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr2_-_24661477 | 1.09 |
ENSDART00000078975
ENSDART00000155677 |
trnau1apb
|
tRNA selenocysteine 1 associated protein 1b |
| chr19_+_20194656 | 1.09 |
ENSDART00000169074
|
hoxa4a
|
homeobox A4a |
| chr19_-_9793494 | 1.08 |
ENSDART00000134816
|
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr16_+_6693574 | 1.07 |
|
|
|
| chr22_+_38028979 | 1.07 |
ENSDART00000165078
ENSDART00000159906 |
CU659306.1
|
ENSDARG00000100625 |
| chr3_+_6557843 | 1.03 |
ENSDART00000169325
|
nup85
|
nucleoporin 85 |
| chr6_-_39311594 | 0.99 |
|
|
|
| chr10_-_36738730 | 0.97 |
ENSDART00000139097
|
dnajb13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr21_+_20347283 | 0.96 |
ENSDART00000026430
|
hspb11
|
heat shock protein, alpha-crystallin-related, b11 |
| chr10_+_35479226 | 0.96 |
ENSDART00000147303
|
hhla2a.1
|
HERV-H LTR-associating 2a, tandem duplicate 1 |
| chr1_-_30298698 | 0.96 |
|
|
|
| chr9_-_47814390 | 0.95 |
|
|
|
| chr6_-_24293008 | 0.95 |
ENSDART00000168355
|
brdt
|
bromodomain, testis-specific |
| chr15_-_177068 | 0.94 |
ENSDART00000115233
|
VSTM5
|
V-set and transmembrane domain containing 5 |
| chr16_-_49282668 | 0.93 |
|
|
|
| chr2_-_35120439 | 0.91 |
ENSDART00000111730
|
pappa2
|
pappalysin 2 |
| chr19_-_9793444 | 0.90 |
ENSDART00000134816
|
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr8_+_28433869 | 0.85 |
ENSDART00000062716
|
slc52a3
|
solute carrier family 52 (riboflavin transporter), member 3 |
| chr11_-_16067646 | 0.85 |
|
|
|
| chr10_-_42155790 | 0.84 |
|
|
|
| chr4_-_30124515 | 0.83 |
|
|
|
| chr21_-_661222 | 0.83 |
|
|
|
| chr25_+_21735567 | 0.82 |
ENSDART00000148299
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr10_+_15398004 | 0.82 |
|
|
|
| chr3_-_32409303 | 0.80 |
ENSDART00000151476
|
rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr17_-_42778156 | 0.77 |
ENSDART00000140549
|
nkx2.2a
|
NK2 homeobox 2a |
| chr10_+_29965951 | 0.76 |
ENSDART00000116893
|
SNORD14
|
Small nucleolar RNA SNORD14 |
| chr14_+_49056502 | 0.76 |
ENSDART00000124773
|
ppid
|
peptidylprolyl isomerase D |
| chr13_+_49154065 | 0.73 |
|
|
|
| chr25_-_14337158 | 0.72 |
ENSDART00000103957
|
exoc3l1
|
exocyst complex component 3-like 1 |
| chr25_+_19901741 | 0.67 |
ENSDART00000026401
|
ENSDARG00000004577
|
ENSDARG00000004577 |
| chr1_-_25369419 | 0.64 |
ENSDART00000168640
|
pdcd4a
|
programmed cell death 4a |
| chr16_-_6881218 | 0.64 |
ENSDART00000149070
|
mbpb
|
myelin basic protein b |
| chr22_-_570067 | 0.63 |
ENSDART00000145983
|
cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr23_+_35976073 | 0.63 |
ENSDART00000133760
|
hoxc11a
|
homeobox C11a |
| chr23_+_25005457 | 0.60 |
ENSDART00000136162
|
klhl21
|
kelch-like family member 21 |
| chr25_+_29999307 | 0.59 |
ENSDART00000003346
|
pdcd2l
|
programmed cell death 2-like |
| chr10_+_17978994 | 0.58 |
ENSDART00000146489
|
prkab1a
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit, a |
| chr21_-_25719352 | 0.56 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr8_+_28419432 | 0.55 |
ENSDART00000062706
|
tmem189
|
transmembrane protein 189 |
| chr6_+_19623990 | 0.54 |
ENSDART00000057223
|
rhebl1
|
Ras homolog enriched in brain like 1 |
| chr24_-_20987184 | 0.54 |
ENSDART00000010126
|
zdhhc23b
|
zinc finger, DHHC-type containing 23b |
| chr6_+_60152913 | 0.53 |
|
|
|
| chr17_+_22050144 | 0.51 |
|
|
|
| chr9_-_18727454 | 0.51 |
ENSDART00000143165
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr20_+_19879839 | 0.49 |
ENSDART00000132576
|
BX510657.1
|
ENSDARG00000093899 |
| chr7_+_21006803 | 0.49 |
ENSDART00000052942
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr10_+_29964045 | 0.49 |
ENSDART00000118838
|
SNORD14
|
Small nucleolar RNA SNORD14 |
| chr19_-_18315867 | 0.47 |
ENSDART00000151133
|
top2b
|
topoisomerase (DNA) II beta |
| chr18_+_8388897 | 0.47 |
ENSDART00000083421
|
cpt1b
|
carnitine palmitoyltransferase 1B (muscle) |
| chr7_-_6334156 | 0.45 |
ENSDART00000081359
|
zgc:110425
|
zgc:110425 |
| chr22_+_10411257 | 0.45 |
ENSDART00000064805
|
cenpp
|
centromere protein P |
| chr23_+_17946933 | 0.42 |
ENSDART00000117743
|
SNORD59
|
Small nucleolar RNA SNORD59 |
| chr9_+_6609374 | 0.41 |
ENSDART00000122279
|
fhl2a
|
four and a half LIM domains 2a |
| chr1_-_58281796 | 0.41 |
ENSDART00000175765
|
CABZ01109030.1
|
ENSDARG00000108798 |
| chr1_-_55072064 | 0.40 |
ENSDART00000142244
|
dnajb1b
|
DnaJ (Hsp40) homolog, subfamily B, member 1b |
| chr16_-_22394481 | 0.39 |
|
|
|
| chr17_-_11264425 | 0.39 |
ENSDART00000151847
|
arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr24_+_32635139 | 0.38 |
ENSDART00000175986
|
CU138525.1
|
ENSDARG00000105889 |
| chr18_+_14509074 | 0.37 |
ENSDART00000146472
|
kcng4a
|
potassium voltage-gated channel, subfamily G, member 4a |
| chr19_+_5054263 | 0.37 |
|
|
|
| chr15_-_22212082 | 0.37 |
ENSDART00000159116
|
scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr10_+_29963998 | 0.36 |
ENSDART00000118838
|
SNORD14
|
Small nucleolar RNA SNORD14 |
| chr10_-_40904685 | 0.32 |
|
|
|
| chr1_-_30298665 | 0.30 |
|
|
|
| chr13_+_24703728 | 0.26 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr2_-_11720434 | 0.25 |
ENSDART00000019392
|
sdr16c5a
|
short chain dehydrogenase/reductase family 16C, member 5a |
| chr14_-_32485197 | 0.24 |
|
|
|
| chr9_-_47814321 | 0.24 |
|
|
|
| chr6_-_39312114 | 0.24 |
|
|
|
| chr11_+_29753292 | 0.23 |
|
|
|
| chr12_+_19745010 | 0.23 |
ENSDART00000045609
|
cpped1
|
calcineurin-like phosphoesterase domain containing 1 |
| chr16_-_32267264 | 0.22 |
ENSDART00000112348
|
fam26e.1
|
family with sequence similarity 26, member E, tandem duplicate 1 |
| chr10_+_29964164 | 0.22 |
ENSDART00000118838
|
SNORD14
|
Small nucleolar RNA SNORD14 |
| chr8_-_23239664 | 0.22 |
|
|
|
| chr7_-_23492042 | 0.22 |
|
|
|
| chr12_+_44829938 | 0.21 |
ENSDART00000098932
|
wbp2
|
WW domain binding protein 2 |
| chr25_-_17282337 | 0.19 |
ENSDART00000064586
|
cyp2x7
|
cytochrome P450, family 2, subfamily X, polypeptide 7 |
| chr3_-_53467536 | 0.19 |
ENSDART00000032788
|
rdh8a
|
retinol dehydrogenase 8a |
| chr7_+_31066952 | 0.17 |
|
|
|
| chr6_-_40700033 | 0.16 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr11_-_35778578 | 0.14 |
ENSDART00000112684
|
setmar
|
SET domain and mariner transposase fusion gene |
| chr1_-_686007 | 0.13 |
ENSDART00000165894
|
adamts1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr6_+_19623749 | 0.13 |
ENSDART00000145564
|
rhebl1
|
Ras homolog enriched in brain like 1 |
| chr7_-_6334242 | 0.13 |
ENSDART00000081359
|
zgc:110425
|
zgc:110425 |
| chr5_-_55745987 | 0.13 |
ENSDART00000083079
|
acaca
|
acetyl-CoA carboxylase alpha |
| chr22_+_21996941 | 0.09 |
ENSDART00000149586
|
gna15.1
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 1 |
| chr13_-_9543393 | 0.07 |
ENSDART00000041609
|
tm9sf3
|
transmembrane 9 superfamily member 3 |
| chr8_-_13327150 | 0.07 |
ENSDART00000063863
|
ccdc124
|
coiled-coil domain containing 124 |
| chr5_+_31211445 | 0.07 |
ENSDART00000023463
|
uap1l1
|
UDP-N-acetylglucosamine pyrophosphorylase 1, like 1 |
| chr4_+_18971779 | 0.06 |
ENSDART00000159160
|
impdh1b
|
IMP (inosine 5'-monophosphate) dehydrogenase 1b |
| chr23_+_17946764 | 0.03 |
ENSDART00000117743
|
SNORD59
|
Small nucleolar RNA SNORD59 |
| chr5_-_24580478 | 0.01 |
ENSDART00000138209
|
dph7
|
diphthamide biosynthesis 7 |
| chr2_+_41973443 | 0.01 |
|
|
|
| chr4_-_12930797 | 0.00 |
ENSDART00000108552
|
lemd3
|
LEM domain containing 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 10.1 | GO:0045428 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 1.2 | 3.6 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 1.0 | 6.0 | GO:0071326 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.9 | 3.8 | GO:1900048 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.8 | 3.8 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.6 | 2.4 | GO:0010660 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.5 | 8.8 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.5 | 3.4 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.4 | 1.7 | GO:1903726 | negative regulation of phospholipid metabolic process(GO:1903726) |
| 0.3 | 4.2 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.3 | 2.9 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.3 | 2.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.3 | 3.0 | GO:0042026 | protein refolding(GO:0042026) |
| 0.3 | 4.3 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.3 | 0.8 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.2 | 2.7 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.2 | 1.5 | GO:0008210 | estrogen biosynthetic process(GO:0006703) estrogen metabolic process(GO:0008210) |
| 0.2 | 4.5 | GO:0044259 | collagen metabolic process(GO:0032963) multicellular organismal macromolecule metabolic process(GO:0044259) |
| 0.2 | 2.3 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.2 | 2.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 0.4 | GO:0086091 | regulation of heart rate by cardiac conduction(GO:0086091) |
| 0.1 | 0.7 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 1.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.8 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 0.8 | GO:0006599 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 6.8 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 0.6 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.1 | 2.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 2.1 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 0.1 | 1.3 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.1 | 1.3 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 1.2 | GO:0072576 | liver morphogenesis(GO:0072576) |
| 0.1 | 0.4 | GO:0034080 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 0.6 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.1 | 1.0 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.1 | 2.3 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 1.1 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 1.6 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 5.3 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.1 | 0.6 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 1.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.9 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.1 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.0 | 2.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.2 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.5 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 3.8 | GO:0003407 | neural retina development(GO:0003407) |
| 0.0 | 0.7 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.6 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 1.6 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 2.8 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.9 | GO:0008593 | regulation of Notch signaling pathway(GO:0008593) |
| 0.0 | 0.2 | GO:0045815 | positive regulation of gene expression, epigenetic(GO:0045815) |
| 0.0 | 3.2 | GO:0007411 | axon guidance(GO:0007411) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.3 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.9 | 10.8 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.6 | 2.9 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.5 | 3.8 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.4 | 3.8 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 1.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 1.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.4 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 3.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 0.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 2.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 8.8 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 2.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 2.1 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 2.2 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.4 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 1.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 6.1 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 2.3 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 3.0 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.5 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.9 | 2.6 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.6 | 2.4 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.6 | 2.9 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.5 | 2.2 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.4 | 2.1 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.4 | 1.1 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.4 | 1.5 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.3 | 2.7 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.3 | 1.2 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.2 | 6.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 3.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 2.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.2 | 2.4 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.2 | 2.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.2 | 2.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 6.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 1.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 1.3 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 0.8 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 5.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 1.6 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 3.0 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 1.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.5 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 8.8 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.1 | 3.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 5.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.6 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.9 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.1 | 5.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 0.8 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 2.7 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) ligase activity, forming carbon-carbon bonds(GO:0016885) |
| 0.0 | 4.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.5 | 6.8 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.3 | 5.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 2.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 0.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 5.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 3.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 2.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.4 | 2.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.4 | 3.0 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.4 | 4.1 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 5.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 0.6 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 5.7 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 0.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.1 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |