Project
DANIO-CODE
Navigation
Downloads
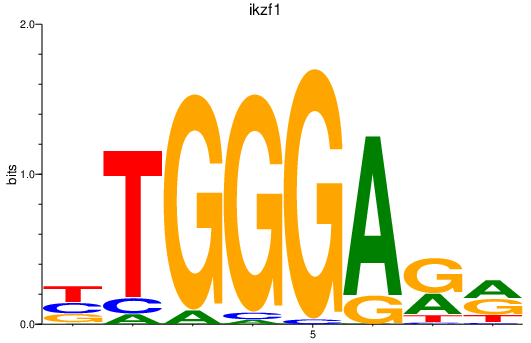
Results for ikzf1
Z-value: 0.82
Transcription factors associated with ikzf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ikzf1
|
ENSDARG00000013539 | IKAROS family zinc finger 1 (Ikaros) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ikzf1 | dr10_dc_chr13_-_15863571_15863574 | 0.24 | 3.7e-01 | Click! |
Activity profile of ikzf1 motif
Sorted Z-values of ikzf1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ikzf1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_3264630 | 1.77 |
ENSDART00000170992
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr15_-_2677200 | 1.71 |
ENSDART00000063320
|
cldne
|
claudin e |
| chr20_+_4567653 | 1.44 |
|
|
|
| chr1_-_50066633 | 1.36 |
|
|
|
| chr11_+_6446302 | 1.21 |
ENSDART00000140707
ENSDART00000036939 |
gadd45ba
|
growth arrest and DNA-damage-inducible, beta a |
| chr18_+_40364675 | 1.16 |
ENSDART00000098791
ENSDART00000049171 |
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr1_+_29054033 | 1.16 |
ENSDART00000054066
|
zic2b
|
zic family member 2 (odd-paired homolog, Drosophila) b |
| chr11_+_7139675 | 1.16 |
ENSDART00000155864
|
CU929070.1
|
ENSDARG00000097452 |
| chr11_-_27706441 | 1.15 |
|
|
|
| chr22_-_10005524 | 1.08 |
ENSDART00000160744
|
BX324216.1
|
ENSDARG00000086839 |
| chr12_+_31558667 | 1.04 |
ENSDART00000152971
|
dnmbp
|
dynamin binding protein |
| chr19_+_17832515 | 1.02 |
ENSDART00000149045
|
klf2b
|
Kruppel-like factor 2b |
| chr24_-_35811390 | 1.02 |
ENSDART00000167990
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr5_-_70948223 | 0.95 |
ENSDART00000013404
|
ak1
|
adenylate kinase 1 |
| chr10_+_33935945 | 0.95 |
|
|
|
| chr15_-_2689005 | 0.87 |
ENSDART00000063325
|
cldnf
|
claudin f |
| chr15_-_20297270 | 0.84 |
ENSDART00000123910
|
ppp1r14ab
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Ab |
| chr5_+_30141562 | 0.84 |
ENSDART00000098246
|
ftr83
|
finTRIM family, member 83 |
| chr24_-_8589270 | 0.78 |
ENSDART00000082346
|
tfap2a
|
transcription factor AP-2 alpha |
| chr15_+_42617510 | 0.77 |
|
|
|
| chr7_-_24201833 | 0.76 |
ENSDART00000121684
|
nat8l
|
N-acetyltransferase 8-like |
| chr14_-_40454194 | 0.73 |
ENSDART00000166621
|
ef1
|
E74-like factor 1 (ets domain transcription factor) |
| chr11_-_29316162 | 0.72 |
ENSDART00000163958
|
arhgef10la
|
Rho guanine nucleotide exchange factor (GEF) 10-like a |
| chr20_+_4567603 | 0.71 |
|
|
|
| chr13_-_36265504 | 0.70 |
ENSDART00000140243
|
actn1
|
actinin, alpha 1 |
| chr23_-_30119058 | 0.69 |
ENSDART00000103480
|
ccdc187
|
coiled-coil domain containing 187 |
| chr16_+_29558320 | 0.67 |
ENSDART00000011497
|
ctsk
|
cathepsin K |
| chr13_-_13163676 | 0.65 |
ENSDART00000125883
ENSDART00000013534 |
fgfr3
|
fibroblast growth factor receptor 3 |
| KN149990v1_+_7315 | 0.63 |
|
|
|
| chr19_+_5375413 | 0.63 |
ENSDART00000141237
|
si:dkeyp-113d7.10
|
si:dkeyp-113d7.10 |
| chr18_-_35415824 | 0.61 |
ENSDART00000098297
|
CABZ01041002.1
|
ENSDARG00000068104 |
| chr24_+_16005004 | 0.61 |
ENSDART00000163086
ENSDART00000152087 |
si:dkey-118j18.2
|
si:dkey-118j18.2 |
| chr1_-_30299017 | 0.60 |
|
|
|
| chr24_+_38783264 | 0.59 |
ENSDART00000154214
|
si:ch73-70c5.1
|
si:ch73-70c5.1 |
| chr24_-_2830194 | 0.58 |
ENSDART00000164913
|
si:ch211-152c8.5
|
si:ch211-152c8.5 |
| chr16_+_46728964 | 0.58 |
ENSDART00000163571
|
rab25b
|
RAB25, member RAS oncogene family b |
| chr23_+_42381296 | 0.58 |
ENSDART00000158684
ENSDART00000159985 ENSDART00000172144 |
cyp2aa11
|
cytochrome P450, family 2, subfamily AA, polypeptide 11 |
| chr23_-_6831711 | 0.57 |
ENSDART00000125393
|
FP102169.1
|
ENSDARG00000089210 |
| chr25_-_2485276 | 0.55 |
ENSDART00000154889
ENSDART00000155027 |
BX957346.1
|
ENSDARG00000096850 |
| chr17_-_14868764 | 0.55 |
ENSDART00000115064
|
nid2a
|
nidogen 2a (osteonidogen) |
| chr13_-_21529695 | 0.54 |
ENSDART00000100925
|
mxtx1
|
mix-type homeobox gene 1 |
| chr2_+_55888093 | 0.54 |
ENSDART00000146160
|
loxl5b
|
lysyl oxidase-like 5b |
| chr3_+_34019696 | 0.53 |
ENSDART00000006091
|
carm1
|
coactivator-associated arginine methyltransferase 1 |
| chr11_+_18862603 | 0.53 |
|
|
|
| chr20_-_54742983 | 0.52 |
|
|
|
| chr15_-_33669717 | 0.52 |
ENSDART00000161151
|
stard13b
|
StAR-related lipid transfer (START) domain containing 13b |
| chr2_-_58111727 | 0.52 |
ENSDART00000004431
ENSDART00000166845 ENSDART00000163999 |
epb41l3b
|
erythrocyte membrane protein band 4.1-like 3b |
| chr7_-_57796486 | 0.52 |
ENSDART00000043984
|
ank2b
|
ankyrin 2b, neuronal |
| chr12_+_29994735 | 0.51 |
ENSDART00000042572
ENSDART00000153025 |
ablim1b
|
actin binding LIM protein 1b |
| chr21_+_30434147 | 0.51 |
ENSDART00000147375
|
snx12
|
sorting nexin 12 |
| chr3_+_30790513 | 0.50 |
ENSDART00000130422
|
cldni
|
claudin i |
| chr11_+_5745644 | 0.49 |
ENSDART00000179139
|
arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr22_-_15678308 | 0.48 |
ENSDART00000130238
|
safb
|
scaffold attachment factor B |
| chr17_-_48623315 | 0.48 |
ENSDART00000030934
|
kcnk5a
|
potassium channel, subfamily K, member 5a |
| chr13_-_39033893 | 0.48 |
ENSDART00000045434
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr5_+_58017820 | 0.47 |
ENSDART00000083015
|
ccdc84
|
coiled-coil domain containing 84 |
| chr6_+_39373026 | 0.47 |
ENSDART00000157165
|
si:dkey-195m11.8
|
si:dkey-195m11.8 |
| chr19_-_28546417 | 0.47 |
ENSDART00000130922
ENSDART00000079114 |
irx1b
|
iroquois homeobox 1b |
| chr4_-_7721419 | 0.46 |
|
|
|
| chr2_-_816669 | 0.46 |
ENSDART00000122732
|
foxc1a
|
forkhead box C1a |
| chr22_+_15871952 | 0.42 |
ENSDART00000062587
|
klf2a
|
Kruppel-like factor 2a |
| chr21_+_25633278 | 0.42 |
ENSDART00000101203
|
sumf2
|
sulfatase modifying factor 2 |
| chr12_-_28840796 | 0.41 |
|
|
|
| chr7_-_38521262 | 0.41 |
ENSDART00000157416
|
si:dkey-23n7.10
|
si:dkey-23n7.10 |
| chr3_+_18779544 | 0.41 |
|
|
|
| chr5_-_25866099 | 0.41 |
ENSDART00000144035
|
arvcfb
|
armadillo repeat gene deleted in velocardiofacial syndrome b |
| chr18_+_17548213 | 0.40 |
ENSDART00000144960
|
nup93
|
nucleoporin 93 |
| chr2_+_42874975 | 0.40 |
ENSDART00000075392
|
basp1
|
brain abundant, membrane attached signal protein 1 |
| chr17_+_33462782 | 0.39 |
ENSDART00000140805
|
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr13_-_13163801 | 0.39 |
ENSDART00000156968
|
fgfr3
|
fibroblast growth factor receptor 3 |
| chr16_+_39396358 | 0.39 |
|
|
|
| chr11_-_4216204 | 0.38 |
ENSDART00000121716
|
ENSDARG00000086300
|
ENSDARG00000086300 |
| chr10_+_34377959 | 0.37 |
|
|
|
| chr8_-_15144533 | 0.37 |
ENSDART00000138855
|
bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr9_-_30744878 | 0.37 |
ENSDART00000079222
|
chaf1b
|
chromatin assembly factor 1, subunit B |
| chr18_-_14891913 | 0.37 |
ENSDART00000018502
|
mapk12a
|
mitogen-activated protein kinase 12a |
| chr13_+_49527181 | 0.37 |
ENSDART00000176643
|
CABZ01084653.1
|
ENSDARG00000106801 |
| chr10_+_32007448 | 0.36 |
ENSDART00000019416
|
lhfp
|
lipoma HMGIC fusion partner |
| chr19_-_23037220 | 0.36 |
ENSDART00000090669
|
pleca
|
plectin a |
| chr9_-_21677993 | 0.35 |
ENSDART00000121939
ENSDART00000080404 |
mphosph8
|
M-phase phosphoprotein 8 |
| chr16_+_53238110 | 0.35 |
ENSDART00000102170
|
CABZ01053976.1
|
ENSDARG00000069929 |
| chr16_+_26892979 | 0.35 |
ENSDART00000048036
|
gem
|
GTP binding protein overexpressed in skeletal muscle |
| chr23_-_30861356 | 0.35 |
ENSDART00000114628
|
myt1a
|
myelin transcription factor 1a |
| chr16_+_5256773 | 0.35 |
ENSDART00000012053
|
elovl4a
|
ELOVL fatty acid elongase 4a |
| chr3_+_31793328 | 0.34 |
ENSDART00000127330
ENSDART00000126773 ENSDART00000055279 |
snrnp70
|
small nuclear ribonucleoprotein 70 (U1) |
| chr5_-_67580755 | 0.34 |
ENSDART00000169350
|
ENSDARG00000098655
|
ENSDARG00000098655 |
| chr3_+_3955069 | 0.34 |
ENSDART00000092393
|
plbd1
|
phospholipase B domain containing 1 |
| chr21_+_18295124 | 0.34 |
ENSDART00000167511
|
ENSDARG00000104101
|
ENSDARG00000104101 |
| chr1_+_8605984 | 0.34 |
ENSDART00000055009
|
uncx4.1
|
Unc4.1 homeobox (C. elegans) |
| chr5_+_31745031 | 0.34 |
ENSDART00000147132
|
c9
|
complement component 9 |
| chr12_+_24994481 | 0.33 |
ENSDART00000170466
|
gch2
|
GTP cyclohydrolase 2 |
| chr16_+_7874573 | 0.33 |
|
|
|
| chr2_+_2583068 | 0.33 |
|
|
|
| chr3_+_52901602 | 0.33 |
ENSDART00000114343
|
brd4
|
bromodomain containing 4 |
| chr3_-_19913881 | 0.33 |
ENSDART00000126915
|
ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr7_-_24119645 | 0.32 |
ENSDART00000141769
|
ptgr1
|
prostaglandin reductase 1 |
| chr12_-_37921731 | 0.32 |
|
|
|
| chr25_+_18467217 | 0.32 |
ENSDART00000170841
|
cav1
|
caveolin 1 |
| chr14_+_21999898 | 0.31 |
|
|
|
| chr11_+_12802775 | 0.31 |
ENSDART00000037474
|
zgpat
|
zinc finger, CCCH-type with G patch domain |
| chr24_-_8589388 | 0.31 |
ENSDART00000082346
|
tfap2a
|
transcription factor AP-2 alpha |
| chr1_-_53746151 | 0.31 |
ENSDART00000179565
|
CABZ01043826.1
|
ENSDARG00000108130 |
| chr19_+_43487281 | 0.30 |
|
|
|
| chr2_-_43347158 | 0.30 |
ENSDART00000141087
|
crema
|
cAMP responsive element modulator a |
| chr2_-_10555152 | 0.30 |
ENSDART00000150166
|
gng12a
|
guanine nucleotide binding protein (G protein), gamma 12a |
| chr2_-_32521879 | 0.29 |
ENSDART00000056639
|
faim2a
|
Fas apoptotic inhibitory molecule 2a |
| chr21_-_2143830 | 0.29 |
|
|
|
| chr12_-_5693715 | 0.29 |
ENSDART00000105887
|
dlx4b
|
distal-less homeobox 4b |
| chr2_-_42643045 | 0.29 |
ENSDART00000139929
|
esyt2a
|
extended synaptotagmin-like protein 2a |
| chr2_+_42342148 | 0.29 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr18_+_45680282 | 0.29 |
ENSDART00000109948
|
qser1
|
glutamine and serine rich 1 |
| chr5_-_1325831 | 0.29 |
|
|
|
| chr20_-_34389652 | 0.28 |
ENSDART00000144705
ENSDART00000020389 |
hmcn1
|
hemicentin 1 |
| chr7_-_17460601 | 0.28 |
ENSDART00000149047
|
men1
|
multiple endocrine neoplasia I |
| chr16_+_5470544 | 0.28 |
|
|
|
| chr16_+_24047649 | 0.28 |
|
|
|
| chr16_+_21109486 | 0.28 |
ENSDART00000079383
|
hoxa9b
|
homeobox A9b |
| chr14_-_45887823 | 0.27 |
ENSDART00000178932
ENSDART00000160657 |
cltb
|
clathrin, light chain B |
| chr8_-_23744125 | 0.27 |
ENSDART00000141871
|
INAVA
|
innate immunity activator |
| chr20_-_7303804 | 0.26 |
ENSDART00000100060
|
dsc2l
|
desmocollin 2 like |
| chr13_+_17333810 | 0.26 |
ENSDART00000134181
|
CT025897.1
|
ENSDARG00000093577 |
| chr9_-_21257082 | 0.26 |
ENSDART00000124533
|
tbx15
|
T-box 15 |
| chr13_-_36265476 | 0.26 |
ENSDART00000133740
ENSDART00000100217 |
actn1
|
actinin, alpha 1 |
| chr23_-_26150495 | 0.26 |
ENSDART00000126299
|
gdi1
|
GDP dissociation inhibitor 1 |
| chr6_+_6333766 | 0.26 |
ENSDART00000140827
|
bcl11ab
|
B-cell CLL/lymphoma 11Ab |
| chr6_-_16328987 | 0.25 |
ENSDART00000083305
|
slc19a2
|
solute carrier family 19 (thiamine transporter), member 2 |
| chr6_+_30441419 | 0.25 |
|
|
|
| chr8_-_13026629 | 0.25 |
ENSDART00000129168
|
si:dkey-208b23.5
|
si:dkey-208b23.5 |
| chr12_-_26339002 | 0.25 |
ENSDART00000153214
|
synpo2lb
|
synaptopodin 2-like b |
| chr5_+_65754237 | 0.25 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr2_-_44402486 | 0.25 |
ENSDART00000011188
ENSDART00000093298 |
atp1a2a
|
ATPase, Na+/K+ transporting, alpha 2a polypeptide |
| chr19_+_12663944 | 0.25 |
ENSDART00000151508
|
ldlrad4a
|
low density lipoprotein receptor class A domain containing 4a |
| chr8_+_20456215 | 0.25 |
ENSDART00000036630
|
zgc:101100
|
zgc:101100 |
| chr25_+_11317025 | 0.24 |
|
|
|
| chr18_+_45680493 | 0.24 |
ENSDART00000109948
|
qser1
|
glutamine and serine rich 1 |
| chr24_-_34794463 | 0.24 |
ENSDART00000171009
ENSDART00000170046 |
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr21_+_45629613 | 0.24 |
|
|
|
| chr6_-_8156520 | 0.24 |
ENSDART00000081561
|
ilf3a
|
interleukin enhancer binding factor 3a |
| chr20_-_54200627 | 0.24 |
|
|
|
| chr5_+_39619826 | 0.23 |
ENSDART00000114078
|
si:dkey-193c22.2
|
si:dkey-193c22.2 |
| chr4_-_12915292 | 0.23 |
ENSDART00000067135
|
msrb3
|
methionine sulfoxide reductase B3 |
| chr13_-_37001997 | 0.23 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr16_-_54634679 | 0.23 |
ENSDART00000075275
ENSDART00000115024 |
pklr
|
pyruvate kinase, liver and RBC |
| chr25_+_5922190 | 0.23 |
ENSDART00000128816
|
sv2
|
synaptic vesicle glycoprotein 2 |
| chr18_-_44365869 | 0.23 |
ENSDART00000166935
|
prdm10
|
PR domain containing 10 |
| chr20_+_27613739 | 0.22 |
|
|
|
| chr5_+_36690093 | 0.22 |
ENSDART00000114671
ENSDART00000113642 |
zgc:163098
|
zgc:163098 |
| chr16_-_31835463 | 0.22 |
ENSDART00000148389
|
chd4b
|
chromodomain helicase DNA binding protein 4b |
| chr2_-_1752776 | 0.22 |
ENSDART00000126566
|
slc22a23
|
solute carrier family 22, member 23 |
| chr7_-_58751849 | 0.22 |
ENSDART00000165390
|
chmp5b
|
charged multivesicular body protein 5b |
| chr24_+_40625215 | 0.22 |
|
|
|
| chr18_-_46831863 | 0.22 |
|
|
|
| chr2_-_1753058 | 0.22 |
ENSDART00000126566
|
slc22a23
|
solute carrier family 22, member 23 |
| chr20_+_35156202 | 0.22 |
ENSDART00000165899
ENSDART00000122696 |
hnrnpub
|
heterogeneous nuclear ribonucleoprotein Ub |
| chr16_+_41922320 | 0.22 |
|
|
|
| chr15_-_25582891 | 0.21 |
|
|
|
| chr21_+_38025480 | 0.21 |
ENSDART00000142106
|
klf8
|
Kruppel-like factor 8 |
| chr10_+_38582701 | 0.21 |
ENSDART00000144329
|
acer3
|
alkaline ceramidase 3 |
| chr23_+_20522512 | 0.21 |
ENSDART00000137294
|
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr9_-_52788707 | 0.20 |
|
|
|
| chr9_-_9693002 | 0.20 |
ENSDART00000018228
|
gsk3b
|
glycogen synthase kinase 3 beta |
| chr14_-_23501428 | 0.20 |
|
|
|
| chr23_+_6861489 | 0.20 |
ENSDART00000092131
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr19_-_7372767 | 0.20 |
ENSDART00000137654
|
sdha
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| chr21_+_7844259 | 0.20 |
ENSDART00000027268
|
otpa
|
orthopedia homeobox a |
| chr13_-_7897936 | 0.19 |
ENSDART00000139728
|
si:ch211-250c4.4
|
si:ch211-250c4.4 |
| chr19_-_46396376 | 0.19 |
|
|
|
| chr14_+_16508093 | 0.19 |
ENSDART00000161201
|
limch1b
|
LIM and calponin homology domains 1b |
| chr23_+_1721759 | 0.19 |
ENSDART00000149545
|
tgm1
|
transglutaminase 1, K polypeptide |
| chr4_+_5370319 | 0.19 |
ENSDART00000163797
|
ENSDARG00000014395
|
ENSDARG00000014395 |
| chr15_-_23441268 | 0.19 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr1_-_52869817 | 0.19 |
ENSDART00000162025
ENSDART00000100788 |
commd1
|
copper metabolism (Murr1) domain containing 1 |
| chr14_-_5371581 | 0.18 |
ENSDART00000012116
|
tlx2
|
T-cell leukemia, homeobox 2 |
| chr10_+_4987494 | 0.18 |
ENSDART00000121959
|
si:ch73-234b20.5
|
si:ch73-234b20.5 |
| chr1_-_23603046 | 0.18 |
|
|
|
| chr18_+_20504980 | 0.18 |
ENSDART00000060295
|
rapsn
|
receptor-associated protein of the synapse, 43kD |
| chr25_-_14948922 | 0.18 |
ENSDART00000161165
|
pax6a
|
paired box 6a |
| chr3_-_60522527 | 0.18 |
ENSDART00000166334
|
srsf2b
|
serine/arginine-rich splicing factor 2b |
| chr20_+_51388214 | 0.18 |
ENSDART00000153452
|
hsp90ab1
|
heat shock protein 90, alpha (cytosolic), class B member 1 |
| chr3_+_17465729 | 0.18 |
ENSDART00000034536
|
rab5c
|
RAB5C, member RAS oncogene family |
| chr3_+_29583029 | 0.18 |
|
|
|
| chr13_-_508202 | 0.18 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
| chr20_+_35156072 | 0.18 |
ENSDART00000165899
ENSDART00000122696 |
hnrnpub
|
heterogeneous nuclear ribonucleoprotein Ub |
| chr4_+_292818 | 0.18 |
ENSDART00000103659
|
msgn1
|
mesogenin 1 |
| chr7_-_18751216 | 0.17 |
ENSDART00000157618
|
HACD4
|
3-hydroxyacyl-CoA dehydratase 4 |
| chr6_-_2028814 | 0.17 |
ENSDART00000171265
|
tgm5l
|
transglutaminase 5, like |
| chr11_-_18004011 | 0.17 |
ENSDART00000125800
|
CABZ01112210.1
|
ENSDARG00000088766 |
| chr19_-_7151715 | 0.17 |
ENSDART00000168755
|
tapbp.2
|
TAP binding protein (tapasin), tandem duplicate 2 |
| chr1_-_30298698 | 0.17 |
|
|
|
| chr17_-_14663139 | 0.17 |
ENSDART00000037371
|
ppp1r13ba
|
protein phosphatase 1, regulatory subunit 13Ba |
| chr6_+_14854074 | 0.17 |
ENSDART00000087782
|
mrps9
|
mitochondrial ribosomal protein S9 |
| chr25_+_31746643 | 0.17 |
ENSDART00000162636
|
tjp1b
|
tight junction protein 1b |
| chr20_+_8041008 | 0.17 |
ENSDART00000085668
|
prkaa2
|
protein kinase, AMP-activated, alpha 2 catalytic subunit |
| chr6_-_39767452 | 0.17 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr7_-_28300008 | 0.17 |
|
|
|
| chr6_-_30671802 | 0.17 |
ENSDART00000065215
|
lurap1
|
leucine rich adaptor protein 1 |
| chr10_+_6925975 | 0.16 |
|
|
|
| chr21_-_40389601 | 0.16 |
ENSDART00000003221
|
nsrp1
|
nuclear speckle splicing regulatory protein 1 |
| chr2_-_1753013 | 0.16 |
ENSDART00000126566
|
slc22a23
|
solute carrier family 22, member 23 |
| chr7_-_33791033 | 0.16 |
ENSDART00000052404
|
map2k5
|
mitogen-activated protein kinase kinase 5 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.4 | 1.2 | GO:1904019 | epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) |
| 0.2 | 0.5 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.2 | 0.9 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.2 | 0.5 | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) regulation of somitogenesis(GO:0014807) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.1 | 1.2 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 1.0 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 3.1 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 0.5 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.1 | 0.4 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.1 | 0.5 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.4 | GO:0046931 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.5 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) protein oxidation(GO:0018158) |
| 0.1 | 0.3 | GO:0010084 | specification of organ axis polarity(GO:0010084) specification of axis polarity(GO:0065001) |
| 0.1 | 0.3 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.1 | 0.3 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 | 0.3 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.3 | GO:0006958 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.7 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 1.1 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) iridophore differentiation(GO:0050935) |
| 0.1 | 0.2 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.1 | 0.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.2 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.1 | 0.2 | GO:0048341 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.1 | 0.3 | GO:1990280 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) regulation of mitotic attachment of spindle microtubules to kinetochore(GO:1902423) positive regulation of attachment of mitotic spindle microtubules to kinetochore(GO:1902425) RNA localization to chromatin(GO:1990280) |
| 0.1 | 1.0 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.2 | GO:0060394 | SMAD protein complex assembly(GO:0007183) regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.2 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.2 | GO:1902306 | negative regulation of sodium ion transmembrane transport(GO:1902306) |
| 0.0 | 0.2 | GO:0036363 | transforming growth factor beta activation(GO:0036363) transforming growth factor beta production(GO:0071604) regulation of transforming growth factor beta production(GO:0071634) negative regulation of transforming growth factor beta production(GO:0071635) regulation of transforming growth factor beta activation(GO:1901388) negative regulation of transforming growth factor beta activation(GO:1901389) |
| 0.0 | 0.2 | GO:2001259 | positive regulation of calcium ion transmembrane transporter activity(GO:1901021) positive regulation of cation channel activity(GO:2001259) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.1 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.0 | 0.4 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.3 | GO:0045117 | azole transport(GO:0045117) |
| 0.0 | 0.2 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.1 | GO:0055016 | hypochord development(GO:0055016) |
| 0.0 | 0.3 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.4 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.6 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 0.1 | GO:1904969 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.0 | 0.7 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.9 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 1.5 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.7 | GO:0030032 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.0 | 0.2 | GO:0061615 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.2 | GO:0090109 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.0 | 0.2 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.5 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.4 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 0.7 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.6 | GO:0009612 | response to mechanical stimulus(GO:0009612) |
| 0.0 | 0.2 | GO:0021684 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.1 | GO:0097531 | mast cell chemotaxis(GO:0002551) positive regulation of leukocyte chemotaxis(GO:0002690) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.5 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.2 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.0 | 0.1 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.0 | 0.2 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.6 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.3 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.5 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0032467 | mitotic spindle elongation(GO:0000022) positive regulation of cytokinesis(GO:0032467) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.3 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.4 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.0 | GO:0035093 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0099631 | postsynaptic endocytic zone cytoplasmic component(GO:0099631) |
| 0.1 | 0.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.2 | GO:0098594 | mucin granule(GO:0098594) |
| 0.1 | 1.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.3 | GO:0000803 | sex chromosome(GO:0000803) inactive sex chromosome(GO:0098577) |
| 0.0 | 0.2 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 3.3 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.2 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.7 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.8 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.4 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.3 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.2 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.1 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.6 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.3 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.2 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.1 | 0.3 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.1 | 0.9 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.5 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.2 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.1 | 1.0 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.8 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 1.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.3 | GO:0017130 | TFIIH-class transcription factor binding(GO:0001097) poly(C) RNA binding(GO:0017130) |
| 0.1 | 0.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.4 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 0.2 | GO:0032038 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.3 | GO:1901474 | azole transporter activity(GO:0045118) vitamin transmembrane transporter activity(GO:0090482) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.6 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.3 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.4 | GO:0022840 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.2 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.2 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 1.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.5 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.6 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 0.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.5 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.4 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 0.7 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 1.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.4 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |