Project
DANIO-CODE
Navigation
Downloads
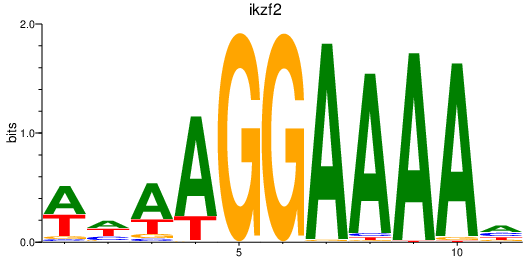
Results for ikzf2
Z-value: 1.62
Transcription factors associated with ikzf2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ikzf2
|
ENSDARG00000069111 | IKAROS family zinc finger 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IKZF2 | dr10_dc_chr9_-_40212643_40212651 | 0.93 | 2.1e-07 | Click! |
Activity profile of ikzf2 motif
Sorted Z-values of ikzf2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ikzf2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_30790513 | 7.00 |
ENSDART00000130422
|
cldni
|
claudin i |
| chr11_+_21749658 | 5.68 |
ENSDART00000161485
|
foxp4
|
forkhead box P4 |
| chr1_+_1541977 | 5.60 |
ENSDART00000048828
|
atp1a1a.4
|
ATPase, Na+/K+ transporting, alpha 1a polypeptide, tandem duplicate 4 |
| chr3_-_48865474 | 5.27 |
ENSDART00000133036
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr16_+_23482744 | 5.08 |
ENSDART00000148961
|
s100a10b
|
S100 calcium binding protein A10b |
| chr7_-_35161302 | 4.60 |
ENSDART00000026712
|
mmp2
|
matrix metallopeptidase 2 |
| chr23_-_39956151 | 3.50 |
ENSDART00000115330
|
ppp1r14c
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr3_+_54876680 | 3.17 |
ENSDART00000111585
|
hbbe1.3
|
hemoglobin, beta embryonic 1.3 |
| chr5_+_8935212 | 3.11 |
|
|
|
| chr24_-_23175007 | 3.09 |
ENSDART00000112256
|
zfhx4
|
zinc finger homeobox 4 |
| chr13_+_24531753 | 2.84 |
ENSDART00000014176
|
msx3
|
muscle segment homeobox 3 |
| chr7_-_2239498 | 2.80 |
ENSDART00000173892
|
si:cabz01007794.1
|
si:cabz01007794.1 |
| chr18_+_26437604 | 2.79 |
ENSDART00000060245
|
ctsh
|
cathepsin H |
| chr3_-_19942214 | 2.78 |
ENSDART00000029386
|
slc4a1a
|
solute carrier family 4 (anion exchanger), member 1a (Diego blood group) |
| chr14_+_23980348 | 2.69 |
ENSDART00000160984
|
BX005228.1
|
ENSDARG00000105465 |
| chr14_-_42633578 | 2.65 |
ENSDART00000162714
ENSDART00000161521 |
pcdh10b
|
protocadherin 10b |
| chr7_+_25649559 | 2.55 |
ENSDART00000026295
|
arrb2b
|
arrestin, beta 2b |
| chr12_-_9479063 | 2.52 |
ENSDART00000169727
|
si:ch211-207i20.3
|
si:ch211-207i20.3 |
| chr16_+_26900732 | 2.51 |
ENSDART00000042895
|
cdh17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr16_+_19926776 | 2.47 |
ENSDART00000149901
ENSDART00000052927 |
twist1b
|
twist family bHLH transcription factor 1b |
| chr23_-_21520413 | 2.47 |
ENSDART00000044080
ENSDART00000112929 |
her12
|
hairy-related 12 |
| chr2_-_43998886 | 2.45 |
ENSDART00000146493
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| chr20_-_21031504 | 2.44 |
ENSDART00000152726
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr18_+_48433986 | 2.39 |
ENSDART00000076861
|
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr6_+_29800606 | 2.38 |
ENSDART00000017424
|
ptmaa
|
prothymosin, alpha a |
| chr18_+_19659195 | 2.37 |
ENSDART00000100569
|
smad6b
|
SMAD family member 6b |
| chr1_+_8605984 | 2.37 |
ENSDART00000055009
|
uncx4.1
|
Unc4.1 homeobox (C. elegans) |
| chr4_-_24298444 | 2.37 |
ENSDART00000077926
ENSDART00000128368 |
celf2
|
cugbp, Elav-like family member 2 |
| chr17_+_33766838 | 2.31 |
ENSDART00000132294
|
fut8a
|
fucosyltransferase 8a (alpha (1,6) fucosyltransferase) |
| chr3_+_13114740 | 2.29 |
ENSDART00000162724
|
ENSDARG00000074231
|
ENSDARG00000074231 |
| chr2_+_3701942 | 2.26 |
ENSDART00000132572
|
gpt2l
|
glutamic pyruvate transaminase (alanine aminotransferase) 2, like |
| chr16_+_5470544 | 2.26 |
|
|
|
| chr9_+_53736715 | 2.24 |
ENSDART00000003310
|
sox21b
|
SRY (sex determining region Y)-box 21b |
| chr16_+_17705704 | 2.21 |
|
|
|
| chr15_+_19902697 | 2.20 |
ENSDART00000101204
|
alcamb
|
activated leukocyte cell adhesion molecule b |
| chr25_-_255242 | 2.20 |
|
|
|
| chr14_+_21531709 | 2.16 |
ENSDART00000144367
|
ctbp1
|
C-terminal binding protein 1 |
| chr22_-_19986461 | 2.16 |
ENSDART00000093310
|
celf5a
|
cugbp, Elav-like family member 5a |
| chr24_-_23174888 | 2.14 |
ENSDART00000112256
|
zfhx4
|
zinc finger homeobox 4 |
| chr3_-_24925189 | 2.14 |
ENSDART00000156459
|
ep300b
|
E1A binding protein p300 b |
| chr3_+_37565550 | 2.14 |
ENSDART00000151236
|
map3k14a
|
mitogen-activated protein kinase kinase kinase 14a |
| chr1_-_55554518 | 2.12 |
ENSDART00000019573
|
zgc:65894
|
zgc:65894 |
| chr2_-_30071872 | 2.11 |
ENSDART00000056747
|
shhb
|
sonic hedgehog b |
| chr20_-_29148810 | 2.11 |
ENSDART00000140350
|
thbs1b
|
thrombospondin 1b |
| chr12_-_48493654 | 2.10 |
ENSDART00000162603
|
ndufb8
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8 |
| chr2_+_29992879 | 2.09 |
ENSDART00000056748
|
en2b
|
engrailed homeobox 2b |
| chr7_-_33080261 | 2.09 |
ENSDART00000114041
|
anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr23_+_25782195 | 2.08 |
ENSDART00000060059
|
rbms2b
|
RNA binding motif, single stranded interacting protein 2b |
| chr9_-_24183317 | 2.07 |
ENSDART00000140346
|
col6a3
|
collagen, type VI, alpha 3 |
| chr1_+_45063098 | 2.07 |
ENSDART00000084512
|
pkn1a
|
protein kinase N1a |
| chr5_-_33265038 | 2.06 |
ENSDART00000132634
|
BX005445.1
|
ENSDARG00000095064 |
| chr21_-_7277655 | 2.05 |
ENSDART00000056561
|
s100z
|
S100 calcium binding protein Z |
| chr23_-_20837809 | 2.04 |
ENSDART00000089750
|
znf362b
|
zinc finger protein 362b |
| chr15_+_30276801 | 2.03 |
ENSDART00000047248
ENSDART00000123937 |
nlk2
|
nemo-like kinase, type 2 |
| chr8_+_10267246 | 1.94 |
ENSDART00000159312
ENSDART00000160766 |
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr23_+_23730717 | 1.94 |
|
|
|
| chr7_+_36280563 | 1.92 |
ENSDART00000027807
|
chd9
|
chromodomain helicase DNA binding protein 9 |
| chr16_+_29558320 | 1.92 |
ENSDART00000011497
|
ctsk
|
cathepsin K |
| chr21_+_5693372 | 1.89 |
ENSDART00000020603
|
ccng2
|
cyclin G2 |
| chr1_-_5796394 | 1.86 |
ENSDART00000109356
|
klf7a
|
Kruppel-like factor 7a |
| chr7_-_72278552 | 1.85 |
ENSDART00000168532
|
HECTD4
|
HECT domain E3 ubiquitin protein ligase 4 |
| chr23_+_35988395 | 1.85 |
ENSDART00000154825
|
hoxc3a
|
homeo box C3a |
| chr1_+_6995893 | 1.84 |
ENSDART00000163488
|
en1b
|
engrailed homeobox 1b |
| chr5_-_63422783 | 1.83 |
ENSDART00000083684
|
pappab
|
pregnancy-associated plasma protein A, pappalysin 1b |
| chr7_+_39116005 | 1.80 |
ENSDART00000146702
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr22_+_13862110 | 1.80 |
ENSDART00000105711
|
sh3bp4a
|
SH3-domain binding protein 4a |
| chr7_+_58443471 | 1.78 |
ENSDART00000148851
|
rps20
|
ribosomal protein S20 |
| chr6_-_12487617 | 1.77 |
ENSDART00000090174
|
dock9b
|
dedicator of cytokinesis 9b |
| chr14_+_20632593 | 1.77 |
ENSDART00000166366
ENSDART00000106198 |
zgc:66433
|
zgc:66433 |
| chr2_+_21342233 | 1.76 |
ENSDART00000062563
|
rreb1b
|
ras responsive element binding protein 1b |
| chr13_-_47767713 | 1.75 |
ENSDART00000045475
|
itga9
|
integrin, alpha 9 |
| chr6_+_35378839 | 1.74 |
ENSDART00000102483
ENSDART00000133783 |
rgs4
|
regulator of G protein signaling 4 |
| chr5_-_25866099 | 1.74 |
ENSDART00000144035
|
arvcfb
|
armadillo repeat gene deleted in velocardiofacial syndrome b |
| chr19_+_1814994 | 1.74 |
|
|
|
| chr11_-_32461160 | 1.74 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr16_+_33702010 | 1.71 |
ENSDART00000143757
|
fhl3a
|
four and a half LIM domains 3a |
| chr23_-_7740845 | 1.71 |
ENSDART00000172451
|
plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr7_-_46509291 | 1.69 |
ENSDART00000173891
|
tshz3b
|
teashirt zinc finger homeobox 3b |
| chr3_+_29583029 | 1.68 |
|
|
|
| chr23_+_36002332 | 1.65 |
ENSDART00000103139
|
hoxc8a
|
homeobox C8a |
| chr11_-_25962291 | 1.64 |
|
|
|
| chr10_+_9325719 | 1.63 |
ENSDART00000064968
|
rasgef1bb
|
RasGEF domain family, member 1Bb |
| chr17_+_52736192 | 1.63 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr10_+_10428297 | 1.63 |
ENSDART00000179214
|
sardh
|
sarcosine dehydrogenase |
| chr3_+_12432686 | 1.62 |
|
|
|
| chr5_-_68145952 | 1.61 |
ENSDART00000141917
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr6_-_36574848 | 1.60 |
ENSDART00000135413
|
her6
|
hairy-related 6 |
| chr13_-_47767748 | 1.60 |
ENSDART00000045475
|
itga9
|
integrin, alpha 9 |
| chr24_-_9151388 | 1.59 |
ENSDART00000149875
|
tgif1
|
TGFB-induced factor homeobox 1 |
| chr19_-_20614846 | 1.59 |
ENSDART00000155527
|
si:ch211-155k24.9
|
si:ch211-155k24.9 |
| chr8_+_46651079 | 1.57 |
ENSDART00000113803
|
her3
|
hairy-related 3 |
| chr13_+_4536343 | 1.55 |
|
|
|
| chr1_-_40208544 | 1.55 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr21_-_24552672 | 1.54 |
|
|
|
| chr19_-_10324632 | 1.54 |
ENSDART00000148073
|
shisa7b
|
shisa family member 7 |
| chr6_+_58921655 | 1.52 |
ENSDART00000083628
|
ddit3
|
DNA-damage-inducible transcript 3 |
| chr21_-_20674965 | 1.51 |
ENSDART00000065649
|
ENSDARG00000044676
|
ENSDARG00000044676 |
| chr25_+_30472954 | 1.51 |
|
|
|
| chr2_-_21960233 | 1.51 |
ENSDART00000027587
|
ralab
|
v-ral simian leukemia viral oncogene homolog Ab (ras related) |
| chr5_+_34397578 | 1.50 |
ENSDART00000043341
|
foxd1
|
forkhead box D1 |
| chr13_-_23536022 | 1.49 |
|
|
|
| chr2_-_30071815 | 1.48 |
ENSDART00000056747
|
shhb
|
sonic hedgehog b |
| chr3_-_48865399 | 1.47 |
ENSDART00000028610
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr17_+_22082952 | 1.47 |
ENSDART00000047772
|
mal
|
mal, T-cell differentiation protein |
| chr18_+_7632469 | 1.46 |
ENSDART00000136313
|
si:dkeyp-1h4.6
|
si:dkeyp-1h4.6 |
| chr3_+_34064081 | 1.45 |
|
|
|
| chr7_+_20272091 | 1.45 |
ENSDART00000052917
|
slc3a2a
|
solute carrier family 3 (amino acid transporter heavy chain), member 2a |
| chr9_+_56182756 | 1.45 |
ENSDART00000144757
|
mxra5b
|
matrix-remodelling associated 5b |
| chr25_-_13057808 | 1.45 |
ENSDART00000172571
|
smpd3
|
sphingomyelin phosphodiesterase 3, neutral |
| chr8_-_52426509 | 1.45 |
ENSDART00000015081
|
ENSDARG00000011208
|
ENSDARG00000011208 |
| chr11_-_25803101 | 1.44 |
ENSDART00000088888
|
kaznb
|
kazrin, periplakin interacting protein b |
| chr12_+_28252623 | 1.44 |
ENSDART00000066294
|
cdk5r1b
|
cyclin-dependent kinase 5, regulatory subunit 1b (p35) |
| chr4_+_18854068 | 1.43 |
ENSDART00000066977
|
bik
|
BCL2-interacting killer (apoptosis-inducing) |
| chr9_-_23346038 | 1.43 |
ENSDART00000135461
|
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr1_-_51862897 | 1.43 |
ENSDART00000136469
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr1_-_40208469 | 1.42 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr11_-_23151247 | 1.42 |
|
|
|
| chr23_+_25392850 | 1.42 |
|
|
|
| chr13_+_13562712 | 1.42 |
ENSDART00000110509
ENSDART00000169265 |
si:ch211-194c3.5
|
si:ch211-194c3.5 |
| chr4_+_5240888 | 1.41 |
ENSDART00000150391
|
si:ch211-214j24.14
|
si:ch211-214j24.14 |
| chr5_-_21523520 | 1.40 |
ENSDART00000023306
|
asb12a
|
ankyrin repeat and SOCS box-containing 12a |
| chr3_-_12432661 | 1.40 |
|
|
|
| chr7_-_54407681 | 1.39 |
ENSDART00000162795
|
ccnd1
|
cyclin D1 |
| chr23_+_6298911 | 1.38 |
ENSDART00000139795
|
syt2a
|
synaptotagmin IIa |
| chr22_-_24341408 | 1.38 |
|
|
|
| chr9_+_46038777 | 1.37 |
ENSDART00000114814
|
twist2
|
twist2 |
| chr16_+_19731017 | 1.37 |
ENSDART00000139357
|
sp8b
|
sp8 transcription factor b |
| chr11_+_21749918 | 1.35 |
ENSDART00000161485
|
foxp4
|
forkhead box P4 |
| chr24_+_9231693 | 1.34 |
ENSDART00000082422
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr6_+_39362786 | 1.34 |
ENSDART00000151322
|
ENSDARG00000028618
|
ENSDARG00000028618 |
| chr5_+_19924978 | 1.34 |
ENSDART00000141118
|
tmem119a
|
transmembrane protein 119a |
| chr9_-_33518818 | 1.34 |
ENSDART00000140039
|
rpl8
|
ribosomal protein L8 |
| chr4_-_23362106 | 1.33 |
ENSDART00000133644
ENSDART00000009768 |
magi2a
|
membrane associated guanylate kinase, WW and PDZ domain containing 2a |
| chr4_+_4223606 | 1.33 |
|
|
|
| chr2_-_54965656 | 1.33 |
|
|
|
| chr19_+_15538967 | 1.32 |
ENSDART00000171403
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr24_-_11681151 | 1.31 |
ENSDART00000058992
|
gli3
|
GLI family zinc finger 3 |
| chr22_+_34738389 | 1.30 |
ENSDART00000154372
|
hoga1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr24_-_11681037 | 1.30 |
ENSDART00000058992
|
gli3
|
GLI family zinc finger 3 |
| chr4_-_6558919 | 1.29 |
ENSDART00000142087
|
foxp2
|
forkhead box P2 |
| KN150442v1_-_35734 | 1.29 |
|
|
|
| chr8_+_7655655 | 1.29 |
ENSDART00000170184
|
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr12_-_7902815 | 1.28 |
ENSDART00000088100
|
ank3b
|
ankyrin 3b |
| chr25_-_21618526 | 1.28 |
ENSDART00000152011
|
DOCK4 (1 of many)
|
dedicator of cytokinesis 4 |
| chr24_+_14093524 | 1.27 |
|
|
|
| chr23_-_18972097 | 1.25 |
ENSDART00000133826
|
CR847953.1
|
ENSDARG00000057403 |
| chr22_-_16009772 | 1.24 |
|
|
|
| chr14_+_33073643 | 1.23 |
ENSDART00000052780
|
ndufa1
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1 |
| chr16_-_10089440 | 1.23 |
ENSDART00000066372
|
id4
|
inhibitor of DNA binding 4 |
| chr4_+_52697064 | 1.23 |
|
|
|
| chr13_-_16126849 | 1.22 |
ENSDART00000079745
|
zgc:110045
|
zgc:110045 |
| chr23_-_27721250 | 1.22 |
ENSDART00000059974
|
wnt10b
|
wingless-type MMTV integration site family, member 10b |
| chr16_+_5466342 | 1.22 |
ENSDART00000160008
|
plecb
|
plectin b |
| chr21_+_25597814 | 1.22 |
ENSDART00000110705
|
tmem151a
|
transmembrane protein 151A |
| chr7_+_73598802 | 1.21 |
ENSDART00000109720
|
zgc:163061
|
zgc:163061 |
| chr3_+_23587156 | 1.20 |
|
|
|
| chr10_-_36864268 | 1.19 |
ENSDART00000165853
|
dhrs13a.1
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 1 |
| chr7_-_40713381 | 1.19 |
ENSDART00000031700
|
en2a
|
engrailed homeobox 2a |
| chr14_-_14260824 | 1.19 |
ENSDART00000159056
|
si:dkey-27i16.2
|
si:dkey-27i16.2 |
| chr6_+_4000287 | 1.19 |
ENSDART00000111817
|
trim25l
|
tripartite motif containing 25, like |
| chr21_+_5293086 | 1.18 |
|
|
|
| chr19_-_30834919 | 1.17 |
ENSDART00000043554
|
ppp1r10
|
protein phosphatase 1, regulatory subunit 10 |
| chr3_+_23561502 | 1.17 |
ENSDART00000078453
|
hoxb7a
|
homeobox B7a |
| chr1_-_22144014 | 1.17 |
ENSDART00000043556
|
ldb2b
|
LIM domain binding 2b |
| chr12_-_3742062 | 1.16 |
ENSDART00000092983
|
ENSDARG00000063555
|
ENSDARG00000063555 |
| chr9_+_5884066 | 1.15 |
ENSDART00000129117
|
pdzk1
|
PDZ domain containing 1 |
| chr20_+_26803282 | 1.15 |
ENSDART00000077753
|
foxc1b
|
forkhead box C1b |
| chr4_-_16365281 | 1.15 |
ENSDART00000139919
|
lum
|
lumican |
| chr13_+_22134507 | 1.14 |
ENSDART00000060576
|
myoz1a
|
myozenin 1a |
| chr23_-_45266310 | 1.14 |
|
|
|
| chr9_-_8475669 | 1.13 |
ENSDART00000110158
|
irs2b
|
insulin receptor substrate 2b |
| chr7_+_19300351 | 1.12 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr11_+_33972923 | 1.12 |
ENSDART00000172594
|
fam43a
|
family with sequence similarity 43, member A |
| chr9_+_38814714 | 1.11 |
ENSDART00000114728
|
znf148
|
zinc finger protein 148 |
| chr12_-_34786844 | 1.10 |
ENSDART00000027379
|
bicral
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr23_-_26562635 | 1.10 |
ENSDART00000168052
|
magi3a
|
membrane associated guanylate kinase, WW and PDZ domain containing 3a |
| chr3_-_19942332 | 1.10 |
ENSDART00000124326
|
slc4a1a
|
solute carrier family 4 (anion exchanger), member 1a (Diego blood group) |
| chr1_-_23771836 | 1.09 |
ENSDART00000126950
|
sh3d19
|
SH3 domain containing 19 |
| chr19_+_30210743 | 1.08 |
ENSDART00000167803
|
marcksl1b
|
MARCKS-like 1b |
| chr13_-_31516399 | 1.08 |
ENSDART00000172375
ENSDART00000125987 ENSDART00000143903 ENSDART00000158719 |
six4a
|
SIX homeobox 4a |
| chr25_-_13456748 | 1.08 |
ENSDART00000139290
|
ano10b
|
anoctamin 10b |
| chr22_-_19986607 | 1.07 |
ENSDART00000093310
|
celf5a
|
cugbp, Elav-like family member 5a |
| chr19_-_46058954 | 1.07 |
ENSDART00000163504
|
trps1
|
trichorhinophalangeal syndrome I |
| chr22_-_18724510 | 1.07 |
ENSDART00000167466
|
midn
|
midnolin |
| chr23_+_36007936 | 1.06 |
ENSDART00000128533
|
hoxc3a
|
homeo box C3a |
| chr5_-_43686611 | 1.06 |
ENSDART00000146080
|
si:ch73-337l15.2
|
si:ch73-337l15.2 |
| chr4_+_11054808 | 1.06 |
ENSDART00000140362
|
ccdc59
|
coiled-coil domain containing 59 |
| chr12_+_1021520 | 1.06 |
|
|
|
| chr21_-_20728623 | 1.05 |
ENSDART00000135940
ENSDART00000002231 |
ghrb
|
growth hormone receptor b |
| chr23_+_35988426 | 1.05 |
ENSDART00000154825
|
hoxc3a
|
homeo box C3a |
| chr5_-_54026450 | 1.04 |
ENSDART00000159009
|
spag8
|
sperm associated antigen 8 |
| chr14_-_15651090 | 1.04 |
ENSDART00000169197
|
flt4
|
fms-related tyrosine kinase 4 |
| chr16_+_33701759 | 1.04 |
ENSDART00000143757
|
fhl3a
|
four and a half LIM domains 3a |
| chr10_+_39140943 | 1.03 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr6_-_54103765 | 1.03 |
ENSDART00000083880
|
hyal2a
|
hyaluronoglucosaminidase 2a |
| chr16_-_34304851 | 1.03 |
ENSDART00000145485
|
phactr4b
|
phosphatase and actin regulator 4b |
| chr16_+_17706003 | 1.03 |
|
|
|
| chr7_-_54407461 | 1.03 |
ENSDART00000162795
|
ccnd1
|
cyclin D1 |
| chr24_+_16005004 | 1.03 |
ENSDART00000163086
ENSDART00000152087 |
si:dkey-118j18.2
|
si:dkey-118j18.2 |
| chr2_-_30071993 | 1.02 |
ENSDART00000056747
|
shhb
|
sonic hedgehog b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0043393 | G-protein coupled receptor internalization(GO:0002031) regulation of protein binding(GO:0043393) |
| 0.8 | 2.4 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.6 | 5.2 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.5 | 1.5 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.5 | 4.6 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.4 | 1.3 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.4 | 4.6 | GO:0003348 | cardiac endothelial cell differentiation(GO:0003348) endocardial cell differentiation(GO:0060956) |
| 0.4 | 1.6 | GO:1901052 | sarcosine metabolic process(GO:1901052) |
| 0.4 | 1.2 | GO:0035790 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.4 | 1.8 | GO:0031571 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) mitotic G1 DNA damage checkpoint(GO:0031571) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.4 | 0.7 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.3 | 5.9 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.3 | 1.0 | GO:0060855 | selective angioblast sprouting(GO:0035474) venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 0.3 | 2.3 | GO:0036065 | fucosylation(GO:0036065) |
| 0.3 | 4.2 | GO:0021794 | thalamus development(GO:0021794) |
| 0.3 | 4.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.3 | 0.9 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.3 | 3.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.3 | 0.3 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.3 | 7.0 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.3 | 1.5 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 | 1.0 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 | 0.5 | GO:0071632 | optomotor response(GO:0071632) |
| 0.2 | 0.7 | GO:0032475 | otolith formation(GO:0032475) |
| 0.2 | 1.4 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.2 | 3.0 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.2 | 0.9 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.2 | 1.3 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.2 | 0.6 | GO:0051701 | interaction with host(GO:0051701) |
| 0.2 | 5.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.2 | 1.0 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.2 | 1.4 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.2 | 0.7 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.2 | 1.5 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.2 | 0.9 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 0.2 | 2.5 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.2 | 1.0 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.2 | 1.1 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 1.0 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 1.2 | GO:0003307 | regulation of Wnt signaling pathway involved in heart development(GO:0003307) |
| 0.1 | 0.6 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 2.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 1.4 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.3 | GO:0060997 | dendritic spine development(GO:0060996) dendritic spine morphogenesis(GO:0060997) dendritic spine organization(GO:0097061) |
| 0.1 | 0.7 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) negative regulation of lipid storage(GO:0010888) |
| 0.1 | 0.4 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.5 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.1 | 0.4 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 0.1 | 1.0 | GO:0007622 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.1 | 1.3 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.1 | 0.4 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 | 0.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 1.8 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.1 | 0.5 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 0.9 | GO:0030168 | platelet activation(GO:0030168) |
| 0.1 | 0.7 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.9 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 4.1 | GO:0030901 | midbrain development(GO:0030901) |
| 0.1 | 0.6 | GO:0045899 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 1.9 | GO:0009612 | response to mechanical stimulus(GO:0009612) |
| 0.1 | 2.3 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.1 | 3.3 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.1 | 2.0 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.1 | 1.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.6 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 1.7 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 3.3 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 2.1 | GO:0033339 | pectoral fin development(GO:0033339) |
| 0.1 | 0.8 | GO:0042743 | hydrogen peroxide metabolic process(GO:0042743) |
| 0.1 | 0.8 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 2.1 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) negative regulation of blood vessel morphogenesis(GO:2000181) |
| 0.1 | 0.9 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 2.4 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.1 | 2.1 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.1 | 0.9 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.1 | 0.2 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 0.2 | GO:0030307 | positive regulation of cell growth(GO:0030307) |
| 0.1 | 0.2 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.1 | 1.1 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.2 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.6 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.9 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.0 | 0.5 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.0 | 1.3 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.2 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 2.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 9.9 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 0.1 | GO:0072045 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) convergent extension involved in nephron morphogenesis(GO:0072045) |
| 0.0 | 1.0 | GO:0030032 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.0 | 0.9 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 1.6 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 7.0 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.8 | GO:0038034 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.0 | 0.9 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.0 | 1.9 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 1.2 | GO:0045104 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 3.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 8.7 | GO:0050767 | regulation of neurogenesis(GO:0050767) |
| 0.0 | 1.7 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 0.2 | GO:1901381 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.1 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.7 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 0.7 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.4 | GO:1903707 | negative regulation of hemopoiesis(GO:1903707) |
| 0.0 | 0.4 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.8 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 1.9 | GO:0048593 | camera-type eye morphogenesis(GO:0048593) |
| 0.0 | 0.5 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.3 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.6 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.7 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 1.7 | GO:1990778 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 0.9 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 1.2 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.1 | GO:0033077 | B cell differentiation(GO:0030183) T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.2 | GO:0046113 | nucleobase catabolic process(GO:0046113) |
| 0.0 | 1.0 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.0 | 0.3 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.1 | GO:0018196 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.5 | 3.2 | GO:0031838 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 0.4 | 1.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.2 | 0.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 0.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.2 | 0.7 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.2 | 0.7 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.2 | 0.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 0.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 4.0 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 9.9 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 2.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.9 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.5 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 2.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.9 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 3.3 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.6 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 8.5 | GO:0030017 | sarcomere(GO:0030017) |
| 0.1 | 3.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 2.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 3.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 2.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.2 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.3 | GO:1904949 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.0 | 2.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.3 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 16.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.8 | GO:0015935 | small ribosomal subunit(GO:0015935) |
| 0.0 | 0.6 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 1.0 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 4.4 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0045180 | basal part of cell(GO:0045178) basal cortex(GO:0045180) |
| 0.0 | 0.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.5 | GO:0022626 | cytosolic large ribosomal subunit(GO:0022625) cytosolic ribosome(GO:0022626) |
| 0.0 | 0.9 | GO:0030658 | transport vesicle membrane(GO:0030658) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0070699 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.6 | 2.4 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.6 | 5.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.5 | 1.6 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.5 | 1.4 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.5 | 7.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.5 | 4.6 | GO:0005113 | patched binding(GO:0005113) |
| 0.5 | 3.2 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.5 | 1.8 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.4 | 1.1 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.3 | 1.0 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.3 | 4.0 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.3 | 0.9 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.3 | 1.5 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.3 | 1.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.3 | 1.1 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.3 | 3.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 1.7 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 2.3 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.2 | 1.3 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.2 | 0.6 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.2 | 1.0 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.2 | 2.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 0.7 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.2 | 0.5 | GO:0004953 | icosanoid receptor activity(GO:0004953) |
| 0.2 | 1.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.2 | 1.0 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.2 | 0.5 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 0.1 | 1.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 1.8 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.5 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 2.1 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.0 | GO:0008199 | ferroxidase activity(GO:0004322) ferric iron binding(GO:0008199) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.0 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.8 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 7.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 1.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 2.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.7 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 0.6 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.5 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 1.0 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 2.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.8 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 0.9 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 5.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 2.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 3.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 1.0 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.6 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 1.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 4.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.2 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 2.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) molybdenum ion binding(GO:0030151) |
| 0.0 | 59.2 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 1.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 3.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 2.0 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.2 | GO:0019870 | chloride channel regulator activity(GO:0017081) chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.5 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.8 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.2 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 8.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.8 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.2 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) macromolecule transmembrane transporter activity(GO:0022884) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.3 | 3.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.3 | 4.6 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.2 | 1.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 1.9 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 0.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 2.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 3.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 2.8 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 2.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 1.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 5.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 0.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.1 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 2.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.4 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.0 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.4 | 1.9 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.2 | 1.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 1.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 3.2 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 2.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.8 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 0.5 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.1 | 1.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 1.7 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 0.2 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 0.1 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.1 | 3.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 4.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.6 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.9 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 1.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |