Project
DANIO-CODE
Navigation
Downloads
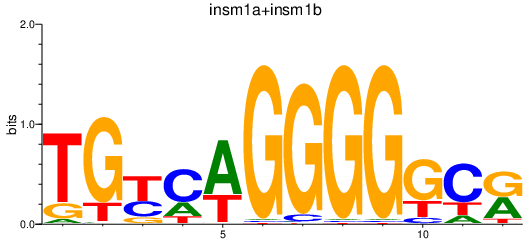
Results for insm1a+insm1b
Z-value: 0.93
Transcription factors associated with insm1a+insm1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
insm1b
|
ENSDARG00000053301 | insulinoma-associated 1b |
|
insm1a
|
ENSDARG00000091756 | insulinoma-associated 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| insm1a | dr10_dc_chr20_-_48677794_48677811 | 0.86 | 2.3e-05 | Click! |
| insm1b | dr10_dc_chr17_+_41475052_41475073 | 0.83 | 6.4e-05 | Click! |
Activity profile of insm1a+insm1b motif
Sorted Z-values of insm1a+insm1b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of insm1a+insm1b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_15774816 | 3.57 |
ENSDART00000090684
|
kremen1
|
kringle containing transmembrane protein 1 |
| chr13_+_371940 | 3.09 |
ENSDART00000013007
|
degs1
|
delta(4)-desaturase, sphingolipid 1 |
| chr3_+_54876680 | 2.93 |
ENSDART00000111585
|
hbbe1.3
|
hemoglobin, beta embryonic 1.3 |
| chr19_+_43039606 | 2.41 |
ENSDART00000010104
|
crtap
|
cartilage associated protein |
| chr22_+_26543146 | 2.39 |
|
|
|
| chr12_+_27370834 | 2.23 |
ENSDART00000105661
|
meox1
|
mesenchyme homeobox 1 |
| chr2_+_24852094 | 2.01 |
ENSDART00000052063
|
rps28
|
ribosomal protein S28 |
| chr4_-_17420378 | 1.95 |
ENSDART00000011943
|
pah
|
phenylalanine hydroxylase |
| chr5_+_42406222 | 1.93 |
ENSDART00000009182
|
aqp3a
|
aquaporin 3a |
| chr12_-_7669319 | 1.92 |
|
|
|
| chr6_-_10593395 | 1.91 |
ENSDART00000020261
|
chrna1
|
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr18_+_7607055 | 1.85 |
ENSDART00000140784
|
CR318588.4
|
ENSDARG00000095556 |
| chr17_+_27383737 | 1.85 |
ENSDART00000156756
|
FP017274.1
|
ENSDARG00000097369 |
| chr5_+_8415025 | 1.75 |
ENSDART00000046440
|
agpat9l
|
1-acylglycerol-3-phosphate O-acyltransferase 9, like |
| chr21_+_5693372 | 1.67 |
ENSDART00000020603
|
ccng2
|
cyclin G2 |
| chr13_+_22173410 | 1.63 |
ENSDART00000173405
|
usp54a
|
ubiquitin specific peptidase 54a |
| chr3_-_45420882 | 1.59 |
ENSDART00000161507
|
zgc:153426
|
zgc:153426 |
| chr7_-_54407681 | 1.52 |
ENSDART00000162795
|
ccnd1
|
cyclin D1 |
| chr23_+_44951868 | 1.51 |
|
|
|
| chr6_-_39311711 | 1.48 |
|
|
|
| chr2_+_33057931 | 1.47 |
|
|
|
| chr1_-_18110990 | 1.45 |
ENSDART00000020970
|
pgm2
|
phosphoglucomutase 2 |
| chr24_-_2918801 | 1.43 |
ENSDART00000164776
|
fam69c
|
family with sequence similarity 69, member C |
| chr3_+_14238188 | 1.38 |
ENSDART00000165452
ENSDART00000171726 |
tmem56b
|
transmembrane protein 56b |
| chr12_-_1544844 | 1.37 |
|
|
|
| chr6_+_13833991 | 1.36 |
|
|
|
| chr6_-_8501230 | 1.28 |
ENSDART00000143956
|
cavin2b
|
caveolae associated protein 2b |
| chr20_-_9212177 | 1.20 |
ENSDART00000064140
|
ylpm1
|
YLP motif containing 1 |
| chr14_+_14262816 | 1.19 |
ENSDART00000164749
|
pcdh20
|
protocadherin 20 |
| chr10_-_15090647 | 1.18 |
ENSDART00000038401
ENSDART00000155674 |
si:ch211-95j8.2
|
si:ch211-95j8.2 |
| chr7_+_22314304 | 1.18 |
ENSDART00000149144
|
chrnb1l
|
cholinergic receptor, nicotinic, beta 1 (muscle) like |
| chr5_+_61753772 | 1.18 |
|
|
|
| chr7_-_6219627 | 1.14 |
ENSDART00000172898
|
HIST2H2AB (1 of many)
|
zgc:165551 |
| chr25_-_23949331 | 1.12 |
ENSDART00000047569
|
igf2b
|
insulin-like growth factor 2b |
| chr19_+_42491631 | 1.11 |
ENSDART00000150949
|
nfyc
|
nuclear transcription factor Y, gamma |
| chr11_-_25803101 | 1.09 |
ENSDART00000088888
|
kaznb
|
kazrin, periplakin interacting protein b |
| chr8_-_14446883 | 1.06 |
ENSDART00000057644
|
lhx4
|
LIM homeobox 4 |
| chr8_-_37358627 | 1.04 |
ENSDART00000009569
|
slc12a5b
|
solute carrier family 12 (potassium/chloride transporter), member 5b |
| chr12_-_4648262 | 1.02 |
ENSDART00000152771
|
si:ch211-255p10.3
|
si:ch211-255p10.3 |
| chr11_+_25382443 | 1.00 |
ENSDART00000033702
|
comtb
|
catechol-O-methyltransferase b |
| chr3_+_17210396 | 0.99 |
ENSDART00000090676
|
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr7_-_54407461 | 0.99 |
ENSDART00000162795
|
ccnd1
|
cyclin D1 |
| chr5_-_15774729 | 0.97 |
ENSDART00000090684
|
kremen1
|
kringle containing transmembrane protein 1 |
| chr7_-_41209891 | 0.97 |
ENSDART00000174300
|
smarcd3b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3b |
| chr1_+_1964902 | 0.96 |
ENSDART00000138396
|
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr4_-_1346961 | 0.96 |
ENSDART00000164623
|
ptn
|
pleiotrophin |
| chr17_-_4160288 | 0.96 |
ENSDART00000153824
|
napba
|
N-ethylmaleimide-sensitive factor attachment protein, beta a |
| chr10_+_21843488 | 0.95 |
ENSDART00000160881
|
pcdh1g30
|
protocadherin 1 gamma 30 |
| chr4_-_25282256 | 0.95 |
ENSDART00000066936
|
tmem110l
|
transmembrane protein 110, like |
| chr11_-_30105047 | 0.92 |
ENSDART00000030794
|
tmem169a
|
transmembrane protein 169a |
| chr7_+_9726412 | 0.91 |
ENSDART00000173155
|
adamts17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr4_-_5589261 | 0.91 |
ENSDART00000017349
|
vegfab
|
vascular endothelial growth factor Ab |
| chr25_+_16505438 | 0.85 |
ENSDART00000008986
|
atp6v1e1a
|
ATPase, H+ transporting, lysosomal, V1 subunit E1a |
| chr14_+_8198019 | 0.83 |
ENSDART00000123364
|
vegfba
|
vascular endothelial growth factor Ba |
| chr8_-_23744125 | 0.78 |
ENSDART00000141871
|
INAVA
|
innate immunity activator |
| chr21_+_11683704 | 0.76 |
ENSDART00000081646
|
glrx
|
glutaredoxin (thioltransferase) |
| chr10_+_20112765 | 0.75 |
ENSDART00000027612
|
xpo7
|
exportin 7 |
| chr21_-_24552672 | 0.75 |
|
|
|
| chr24_+_42133471 | 0.75 |
ENSDART00000170514
|
top1mt
|
topoisomerase (DNA) I, mitochondrial |
| chr8_+_8899112 | 0.74 |
ENSDART00000145970
|
si:dkey-83k24.5
|
si:dkey-83k24.5 |
| chr19_+_10912601 | 0.72 |
ENSDART00000053325
|
tomm40l
|
translocase of outer mitochondrial membrane 40 homolog, like |
| chr18_+_27205666 | 0.71 |
|
|
|
| chr2_-_49060868 | 0.69 |
|
|
|
| chr25_-_34608926 | 0.67 |
ENSDART00000130395
|
hist1h4l
|
histone 1, H4, like |
| chr17_-_52493315 | 0.65 |
ENSDART00000156806
|
rps6kl1
|
ribosomal protein S6 kinase-like 1 |
| chr25_-_13579869 | 0.65 |
ENSDART00000090226
|
znf319b
|
zinc finger protein 319b |
| chr5_-_51184375 | 0.64 |
ENSDART00000163616
|
homer1b
|
homer scaffolding protein 1b |
| chr5_+_50432868 | 0.63 |
ENSDART00000097466
|
fam169aa
|
family with sequence similarity 169, member Aa |
| chr6_-_2028814 | 0.62 |
ENSDART00000171265
|
tgm5l
|
transglutaminase 5, like |
| chr14_+_7868515 | 0.62 |
ENSDART00000167228
|
psd2
|
pleckstrin and Sec7 domain containing 2 |
| chr8_+_6533379 | 0.57 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr3_+_55779906 | 0.56 |
|
|
|
| chr13_+_36554728 | 0.56 |
ENSDART00000136030
|
atp5s
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit s |
| chr10_-_41235132 | 0.56 |
ENSDART00000134851
|
aak1b
|
AP2 associated kinase 1b |
| chr21_+_26576104 | 0.53 |
ENSDART00000142667
|
esrra
|
estrogen-related receptor alpha |
| chr6_-_42143595 | 0.52 |
ENSDART00000085472
|
grm2a
|
glutamate receptor, metabotropic 2a |
| chr13_+_30820707 | 0.50 |
ENSDART00000057469
|
vstm4a
|
V-set and transmembrane domain containing 4a |
| chr7_+_8168009 | 0.50 |
|
|
|
| chr1_+_18639896 | 0.50 |
ENSDART00000078594
|
tyrp1b
|
tyrosinase-related protein 1b |
| chr7_-_24723345 | 0.49 |
ENSDART00000002961
|
rcor2
|
REST corepressor 2 |
| chr4_-_5589227 | 0.49 |
ENSDART00000136229
|
vegfab
|
vascular endothelial growth factor Ab |
| chr16_+_54579976 | 0.48 |
|
|
|
| chr12_-_1545141 | 0.48 |
|
|
|
| chr2_-_43820397 | 0.47 |
ENSDART00000165038
|
FO704635.1
|
ENSDARG00000105141 |
| chr9_-_24220466 | 0.46 |
ENSDART00000021218
|
rpe
|
ribulose-5-phosphate-3-epimerase |
| chr3_-_47294253 | 0.45 |
|
|
|
| chr9_+_51539262 | 0.44 |
|
|
|
| chr6_+_39523953 | 0.43 |
|
|
|
| chr5_+_36332564 | 0.42 |
ENSDART00000037879
|
crx
|
cone-rod homeobox |
| chr19_+_42492029 | 0.40 |
ENSDART00000150949
|
nfyc
|
nuclear transcription factor Y, gamma |
| chr2_-_11879298 | 0.38 |
ENSDART00000136748
|
ENSDARG00000039203
|
ENSDARG00000039203 |
| chr10_-_175160 | 0.38 |
|
|
|
| chr14_+_35029615 | 0.38 |
ENSDART00000144702
|
clint1a
|
clathrin interactor 1a |
| chr24_+_42133272 | 0.38 |
ENSDART00000170514
|
top1mt
|
topoisomerase (DNA) I, mitochondrial |
| chr9_+_21524891 | 0.37 |
|
|
|
| chr11_-_3888456 | 0.36 |
ENSDART00000175310
ENSDART00000082404 |
rpn1
|
ribophorin I |
| chr15_-_19186046 | 0.33 |
ENSDART00000108818
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr1_-_58577305 | 0.32 |
|
|
|
| chr18_+_41266453 | 0.32 |
|
|
|
| chr13_+_30820765 | 0.32 |
ENSDART00000114007
ENSDART00000162254 |
vstm4a
|
V-set and transmembrane domain containing 4a |
| chr2_+_7260634 | 0.31 |
ENSDART00000012119
ENSDART00000153404 |
zgc:110366
|
zgc:110366 |
| chr5_+_30690949 | 0.30 |
|
|
|
| chr5_+_61753841 | 0.29 |
|
|
|
| chr8_+_1016353 | 0.27 |
ENSDART00000149276
|
lzts3b
|
leucine zipper, putative tumor suppressor family member 3b |
| chr10_-_1185742 | 0.26 |
ENSDART00000114261
|
bmpr1bb
|
bone morphogenetic protein receptor, type IBb |
| chr3_-_47294091 | 0.25 |
|
|
|
| chr11_+_18897913 | 0.25 |
ENSDART00000164294
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr23_-_439495 | 0.25 |
ENSDART00000146776
|
tspan2b
|
tetraspanin 2b |
| chr11_-_41487157 | 0.24 |
ENSDART00000173312
|
abhd6a
|
abhydrolase domain containing 6a |
| chr10_+_9361595 | 0.23 |
ENSDART00000136364
|
antxr2b
|
anthrax toxin receptor 2b |
| chr8_+_14921850 | 0.23 |
ENSDART00000138548
|
abl2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr11_+_18897769 | 0.21 |
ENSDART00000163006
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr4_+_74885660 | 0.20 |
ENSDART00000164709
|
zgc:153116
|
zgc:153116 |
| chr7_+_54336324 | 0.19 |
ENSDART00000168474
|
fgf3
|
fibroblast growth factor 3 |
| chr14_-_6096120 | 0.19 |
|
|
|
| chr11_-_282662 | 0.19 |
ENSDART00000168461
|
poc1a
|
POC1 centriolar protein A |
| chr14_-_28261949 | 0.19 |
ENSDART00000126095
|
insb
|
preproinsulin b |
| chr12_+_31388957 | 0.17 |
|
|
|
| chr2_+_24851910 | 0.17 |
ENSDART00000133109
|
rps28
|
ribosomal protein S28 |
| chr8_+_41446054 | 0.16 |
ENSDART00000061144
|
arpc5la
|
actin related protein 2/3 complex, subunit 5-like, a |
| chr11_-_282830 | 0.16 |
ENSDART00000173185
|
poc1a
|
POC1 centriolar protein A |
| KN150487v1_+_15682 | 0.13 |
ENSDART00000166996
|
CABZ01074304.1
|
ENSDARG00000100224 |
| chr7_-_18256512 | 0.13 |
ENSDART00000173539
|
rgs12a
|
regulator of G protein signaling 12a |
| chr8_+_41446169 | 0.12 |
ENSDART00000061144
|
arpc5la
|
actin related protein 2/3 complex, subunit 5-like, a |
| chr18_-_6336677 | 0.09 |
ENSDART00000169844
|
iqsec3b
|
IQ motif and Sec7 domain 3b |
| chr12_-_7669275 | 0.08 |
|
|
|
| chr16_+_14981032 | 0.07 |
|
|
|
| chr20_-_4635674 | 0.06 |
|
|
|
| chr13_-_44687273 | 0.05 |
|
|
|
| chr16_-_10425382 | 0.05 |
ENSDART00000104025
ENSDART00000178832 |
flot1b
|
flotillin 1b |
| chr5_-_2540097 | 0.04 |
|
|
|
| chr5_-_51184313 | 0.03 |
ENSDART00000159166
|
homer1b
|
homer scaffolding protein 1b |
| chr7_+_20829933 | 0.02 |
ENSDART00000173612
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr18_+_18011562 | 0.01 |
ENSDART00000005027
|
ENSDARG00000011498
|
ENSDARG00000011498 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.5 | GO:0048901 | anterior lateral line neuromast development(GO:0048901) anterior lateral line neuromast hair cell differentiation(GO:0048903) |
| 0.5 | 2.5 | GO:1902751 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) mitotic G1 DNA damage checkpoint(GO:0031571) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.4 | 1.9 | GO:1902222 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.4 | 1.9 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.4 | 2.2 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.3 | 2.2 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.3 | 2.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.3 | 1.0 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.2 | 1.0 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.2 | 1.1 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.2 | 2.4 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.2 | 1.8 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.2 | 0.5 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.1 | 0.7 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.5 | GO:0006582 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.1 | 2.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 1.1 | GO:0070665 | positive regulation of mononuclear cell proliferation(GO:0032946) positive regulation of T cell proliferation(GO:0042102) positive regulation of lymphocyte proliferation(GO:0050671) positive regulation of leukocyte proliferation(GO:0070665) |
| 0.1 | 1.0 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 0.8 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 1.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 1.0 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 3.1 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 0.2 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 0.5 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 1.0 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 1.9 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.1 | 0.7 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.4 | GO:0045682 | regulation of epidermis development(GO:0045682) |
| 0.0 | 0.6 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.7 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 1.7 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.4 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.7 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.5 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.0 | 0.0 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.0 | 1.0 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 1.6 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 2.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.8 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 1.6 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 1.2 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.0 | 1.0 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 1.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.4 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.4 | 2.9 | GO:0005833 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 0.4 | 3.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 1.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.2 | 1.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 4.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.5 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 1.0 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.7 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.8 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 1.3 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.0 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.9 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.4 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) glycerol transmembrane transporter activity(GO:0015168) |
| 0.5 | 1.4 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.4 | 2.9 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.4 | 1.9 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.3 | 3.1 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.3 | 1.8 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.3 | 0.8 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.2 | 1.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.2 | 0.5 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.2 | 0.7 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.2 | 1.0 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 1.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 2.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.0 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 0.8 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 1.0 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 1.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.3 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 1.4 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 0.6 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 4.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 1.0 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 1.0 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.7 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.5 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.7 | GO:0022884 | protein transmembrane transporter activity(GO:0008320) macromolecule transmembrane transporter activity(GO:0022884) |
| 0.0 | 0.8 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 3.1 | GO:0016705 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen(GO:0016705) |
| 0.0 | 1.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 2.3 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.6 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 2.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 4.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 1.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.9 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 1.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | REACTOME ACETYLCHOLINE BINDING AND DOWNSTREAM EVENTS | Genes involved in Acetylcholine Binding And Downstream Events |
| 0.2 | 1.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 3.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 2.5 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 2.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 2.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.1 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |