Project
DANIO-CODE
Navigation
Downloads
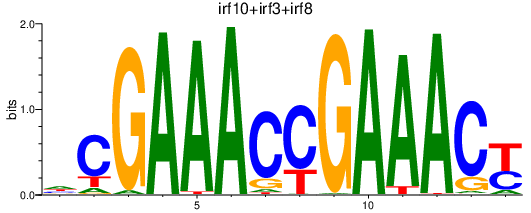
Results for irf10+irf3+irf8
Z-value: 1.71
Transcription factors associated with irf10+irf3+irf8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
irf10
|
ENSDARG00000027658 | interferon regulatory factor 10 |
|
irf8
|
ENSDARG00000056407 | interferon regulatory factor 8 |
|
irf3
|
ENSDARG00000076251 | interferon regulatory factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| irf3 | dr10_dc_chr12_-_4563094_4563107 | 0.67 | 4.4e-03 | Click! |
Activity profile of irf10+irf3+irf8 motif
Sorted Z-values of irf10+irf3+irf8 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of irf10+irf3+irf8
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_43868027 | 7.38 |
ENSDART00000112598
ENSDART00000169576 |
otud4
|
OTU deubiquitinase 4 |
| chr14_-_46980330 | 5.95 |
ENSDART00000043751
ENSDART00000141357 |
macrod1
|
MACRO domain containing 1 |
| chr18_-_38289370 | 4.26 |
ENSDART00000139519
|
caprin1b
|
cell cycle associated protein 1b |
| chr11_+_1558144 | 4.08 |
ENSDART00000154583
|
si:dkey-40c23.2
|
si:dkey-40c23.2 |
| chr9_-_23954640 | 3.99 |
ENSDART00000027212
|
ENSDARG00000006848
|
ENSDARG00000006848 |
| chr13_-_37504883 | 3.95 |
ENSDART00000108855
|
si:dkey-188i13.7
|
si:dkey-188i13.7 |
| chr21_+_22598805 | 3.65 |
ENSDART00000142495
|
BX510940.1
|
ENSDARG00000089088 |
| chr16_+_25370991 | 3.48 |
ENSDART00000154112
|
si:dkey-29h14.10
|
si:dkey-29h14.10 |
| chr22_+_8856370 | 3.45 |
ENSDART00000178799
|
CABZ01046433.1
|
ENSDARG00000109049 |
| chr10_-_35209022 | 3.28 |
ENSDART00000063434
|
ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr18_-_12889296 | 3.24 |
ENSDART00000130343
|
parp12a
|
poly (ADP-ribose) polymerase family, member 12a |
| chr22_-_7431710 | 3.21 |
ENSDART00000170630
|
BX511034.7
|
ENSDARG00000104176 |
| chr24_+_32295105 | 3.09 |
ENSDART00000143570
|
CU207275.1
|
ENSDARG00000092965 |
| chr22_-_7780120 | 3.06 |
ENSDART00000097201
|
si:ch73-44m9.1
|
si:ch73-44m9.1 |
| chr22_-_6062841 | 3.02 |
|
|
|
| chr1_-_55072271 | 2.90 |
ENSDART00000142244
|
dnajb1b
|
DnaJ (Hsp40) homolog, subfamily B, member 1b |
| chr6_+_38898605 | 2.88 |
ENSDART00000029930
|
slc48a1b
|
solute carrier family 48 (heme transporter), member 1b |
| chr5_-_29782745 | 2.86 |
ENSDART00000125381
|
gig2o
|
grass carp reovirus (GCRV)-induced gene 2o |
| chr22_-_8144696 | 2.84 |
ENSDART00000156571
|
si:ch73-44m9.3
|
si:ch73-44m9.3 |
| chr18_-_12889369 | 2.83 |
ENSDART00000130343
|
parp12a
|
poly (ADP-ribose) polymerase family, member 12a |
| chr20_-_22170411 | 2.80 |
ENSDART00000155568
|
BX088688.3
|
ENSDARG00000097598 |
| chr21_+_5027444 | 2.79 |
ENSDART00000139288
|
si:dkey-121h17.7
|
si:dkey-121h17.7 |
| chr9_+_41658126 | 2.77 |
ENSDART00000100265
|
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr4_+_16197 | 2.77 |
ENSDART00000166174
|
ENSDARG00000100660
|
ENSDARG00000100660 |
| chr16_+_25370955 | 2.58 |
ENSDART00000154112
|
si:dkey-29h14.10
|
si:dkey-29h14.10 |
| chr10_-_210594 | 2.55 |
ENSDART00000059476
|
psmg1
|
proteasome (prosome, macropain) assembly chaperone 1 |
| chr16_+_29574449 | 2.50 |
ENSDART00000148450
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr19_+_4135556 | 2.44 |
ENSDART00000162461
|
btr26
|
bloodthirsty-related gene family, member 26 |
| chr9_-_23954690 | 2.44 |
ENSDART00000027212
|
ENSDARG00000006848
|
ENSDARG00000006848 |
| chr25_+_31979838 | 2.33 |
ENSDART00000012600
|
galk2
|
galactokinase 2 |
| chr5_-_66982706 | 2.31 |
ENSDART00000013605
|
zbtb20
|
zinc finger and BTB domain containing 20 |
| chr15_+_20416591 | 2.25 |
ENSDART00000161047
|
il15l
|
interleukin 15, like |
| chr14_-_25636795 | 2.22 |
|
|
|
| chr10_+_35209025 | 2.18 |
ENSDART00000126105
|
nsun5
|
NOP2/Sun domain family, member 5 |
| chr9_+_23954807 | 2.15 |
ENSDART00000145120
|
ENSDARG00000040445
|
ENSDARG00000040445 |
| chr15_+_20054306 | 2.14 |
ENSDART00000155199
|
zgc:112083
|
zgc:112083 |
| chr22_-_8144599 | 2.12 |
ENSDART00000156571
|
si:ch73-44m9.3
|
si:ch73-44m9.3 |
| chr9_-_23954761 | 2.11 |
ENSDART00000027212
|
ENSDARG00000006848
|
ENSDARG00000006848 |
| chr5_+_9579430 | 2.10 |
ENSDART00000109236
|
ENSDARG00000075416
|
ENSDARG00000075416 |
| chr3_+_32530476 | 2.01 |
ENSDART00000171556
|
si:dkey-16l2.16
|
si:dkey-16l2.16 |
| chr11_+_36088518 | 2.01 |
|
|
|
| chr9_+_41657971 | 2.01 |
ENSDART00000132501
|
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr5_-_57053687 | 1.98 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr21_+_5027623 | 1.97 |
ENSDART00000139288
|
si:dkey-121h17.7
|
si:dkey-121h17.7 |
| chr15_-_29229170 | 1.94 |
ENSDART00000138449
|
xaf1
|
XIAP associated factor 1 |
| chr5_-_29782945 | 1.93 |
ENSDART00000125381
|
gig2o
|
grass carp reovirus (GCRV)-induced gene 2o |
| chr15_+_20416518 | 1.92 |
ENSDART00000161047
|
il15l
|
interleukin 15, like |
| chr12_-_19030156 | 1.91 |
ENSDART00000057124
|
tefa
|
thyrotrophic embryonic factor a |
| chr19_-_3947422 | 1.90 |
ENSDART00000172271
|
thrap3b
|
thyroid hormone receptor associated protein 3b |
| chr7_-_69883542 | 1.87 |
|
|
|
| chr24_+_32295150 | 1.86 |
ENSDART00000143570
|
CU207275.1
|
ENSDARG00000092965 |
| chr25_-_25455861 | 1.85 |
ENSDART00000150412
|
irf7
|
interferon regulatory factor 7 |
| chr16_-_9911999 | 1.84 |
ENSDART00000154217
|
tapbpl
|
TAP binding protein (tapasin)-like |
| chr9_-_35824470 | 1.84 |
ENSDART00000140356
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr5_-_38571198 | 1.82 |
ENSDART00000020808
|
paqr3a
|
progestin and adipoQ receptor family member IIIa |
| chr15_-_29229135 | 1.82 |
ENSDART00000138449
|
xaf1
|
XIAP associated factor 1 |
| chr10_-_45383207 | 1.81 |
ENSDART00000169845
|
pargl
|
poly (ADP-ribose) glycohydrolase, like |
| chr17_+_23710004 | 1.80 |
ENSDART00000034913
|
zgc:91976
|
zgc:91976 |
| chr11_-_36088336 | 1.79 |
ENSDART00000141477
|
psma5
|
proteasome subunit alpha 5 |
| chr5_+_22076136 | 1.78 |
ENSDART00000080877
|
tnfsf10l4
|
tumor necrosis factor (ligand) superfamily, member 10 like 4 |
| chr5_-_29782972 | 1.78 |
ENSDART00000125381
|
gig2o
|
grass carp reovirus (GCRV)-induced gene 2o |
| chr9_+_23954669 | 1.71 |
ENSDART00000145120
|
ENSDARG00000040445
|
ENSDARG00000040445 |
| chr7_-_69883581 | 1.69 |
|
|
|
| chr15_-_16447990 | 1.69 |
ENSDART00000154504
|
fam222bb
|
family with sequence similarity 222, member Bb |
| chr12_-_13866903 | 1.69 |
ENSDART00000152400
|
dbf4b
|
DBF4 zinc finger B |
| chr5_-_45358634 | 1.68 |
ENSDART00000135072
|
poc5
|
POC5 centriolar protein homolog (Chlamydomonas) |
| chr4_-_765957 | 1.65 |
ENSDART00000128743
|
tmem214
|
transmembrane protein 214 |
| chr15_-_24653278 | 1.65 |
ENSDART00000123167
|
gemin4
|
gem (nuclear organelle) associated protein 4 |
| chr21_+_5027668 | 1.63 |
ENSDART00000139288
|
si:dkey-121h17.7
|
si:dkey-121h17.7 |
| chr25_-_27222696 | 1.61 |
ENSDART00000123590
|
si:ch211-91p5.3
|
si:ch211-91p5.3 |
| chr15_-_5636737 | 1.61 |
ENSDART00000114410
|
wdr62
|
WD repeat domain 62 |
| chr5_+_45295180 | 1.58 |
ENSDART00000083937
|
polk
|
polymerase (DNA directed) kappa |
| chr22_+_17803309 | 1.58 |
ENSDART00000136016
|
hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr17_+_23709781 | 1.54 |
ENSDART00000034913
|
zgc:91976
|
zgc:91976 |
| chr6_+_46307828 | 1.54 |
ENSDART00000154817
|
si:dkeyp-67f1.1
|
si:dkeyp-67f1.1 |
| chr15_+_14442857 | 1.52 |
ENSDART00000160145
|
si:ch211-11n16.2
|
si:ch211-11n16.2 |
| chr18_+_13280400 | 1.51 |
ENSDART00000080709
|
plcg2
|
phospholipase C, gamma 2 |
| chr12_-_93105 | 1.51 |
ENSDART00000105694
|
adprm
|
ADP-ribose/CDP-alcohol diphosphatase, manganese-dependent |
| chr13_-_49886891 | 1.51 |
ENSDART00000074230
|
pkz
|
protein kinase containing Z-DNA binding domains |
| chr1_-_54570813 | 1.50 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr6_+_12810986 | 1.49 |
ENSDART00000104757
ENSDART00000165896 |
casp8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr15_+_43682831 | 1.49 |
ENSDART00000168589
|
ctsc
|
cathepsin C |
| chr2_-_15380727 | 1.48 |
|
|
|
| chr12_-_13866811 | 1.48 |
ENSDART00000152400
|
dbf4b
|
DBF4 zinc finger B |
| chr7_+_39810607 | 1.47 |
ENSDART00000173559
|
ENSDARG00000036489
|
ENSDARG00000036489 |
| chr22_-_36718623 | 1.47 |
|
|
|
| chr6_-_51811024 | 1.46 |
ENSDART00000031597
|
psmf1
|
proteasome inhibitor subunit 1 |
| chr21_+_22599022 | 1.46 |
ENSDART00000142495
|
BX510940.1
|
ENSDARG00000089088 |
| chr13_+_2345595 | 1.44 |
|
|
|
| chr2_-_6203659 | 1.44 |
ENSDART00000164269
|
prdx1
|
peroxiredoxin 1 |
| chr7_-_667841 | 1.43 |
ENSDART00000160644
|
rela
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr3_+_32530071 | 1.43 |
ENSDART00000165562
|
si:dkey-16l2.16
|
si:dkey-16l2.16 |
| chr21_-_30957555 | 1.43 |
ENSDART00000160224
|
pgap2
|
post-GPI attachment to proteins 2 |
| chr2_+_15380054 | 1.43 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| KN150589v1_-_6303 | 1.42 |
|
|
|
| chr10_-_25366450 | 1.41 |
ENSDART00000123820
|
tmem135
|
transmembrane protein 135 |
| chr3_-_31947991 | 1.40 |
ENSDART00000040900
|
baxb
|
bcl2-associated X protein, b |
| chr3_+_52745606 | 1.40 |
ENSDART00000104683
|
pbx4
|
pre-B-cell leukemia transcription factor 4 |
| chr4_+_17666992 | 1.39 |
ENSDART00000066999
|
ccdc53
|
coiled-coil domain containing 53 |
| chr4_-_26106485 | 1.39 |
ENSDART00000100611
|
si:ch211-244b2.3
|
si:ch211-244b2.3 |
| chr7_-_29956782 | 1.37 |
ENSDART00000173514
|
znf710b
|
zinc finger protein 710b |
| chr22_+_17803347 | 1.37 |
ENSDART00000136016
|
hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr22_-_7887341 | 1.36 |
ENSDART00000097198
|
sc:d217
|
sc:d217 |
| chr2_-_38380883 | 1.36 |
ENSDART00000088026
|
prmt5
|
protein arginine methyltransferase 5 |
| chr4_-_17680826 | 1.35 |
ENSDART00000066997
|
dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr1_-_44227583 | 1.31 |
ENSDART00000134635
|
si:dkey-9i23.15
|
si:dkey-9i23.15 |
| chr2_-_37762426 | 1.29 |
ENSDART00000144807
|
myo9b
|
myosin IXb |
| chr15_-_28852613 | 1.29 |
ENSDART00000152147
|
blmh
|
bleomycin hydrolase |
| chr6_+_41098443 | 1.29 |
ENSDART00000143741
|
fkbp5
|
FK506 binding protein 5 |
| chr11_-_31012985 | 1.29 |
ENSDART00000162605
|
man2b1
|
mannosidase, alpha, class 2B, member 1 |
| chr6_+_48349630 | 1.28 |
ENSDART00000064826
|
mov10a
|
Mov10 RISC complex RNA helicase a |
| chr2_+_16917097 | 1.25 |
ENSDART00000134135
|
si:dkeyp-13a3.10
|
si:dkeyp-13a3.10 |
| chr21_+_5027576 | 1.25 |
ENSDART00000139288
|
si:dkey-121h17.7
|
si:dkey-121h17.7 |
| chr3_+_35997687 | 1.25 |
ENSDART00000058605
|
scpep1
|
serine carboxypeptidase 1 |
| chr9_+_41658313 | 1.25 |
ENSDART00000100265
|
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr19_-_7114660 | 1.24 |
ENSDART00000124094
|
daxx
|
death-domain associated protein |
| chr3_+_12087549 | 1.23 |
|
|
|
| chr21_+_5027911 | 1.22 |
ENSDART00000139288
|
si:dkey-121h17.7
|
si:dkey-121h17.7 |
| chr22_+_24576319 | 1.20 |
ENSDART00000164256
|
ENSDARG00000100186
|
ENSDARG00000100186 |
| chr10_+_9576840 | 1.20 |
ENSDART00000080843
|
ndufa8
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8 |
| chr19_-_7151715 | 1.20 |
ENSDART00000168755
|
tapbp.2
|
TAP binding protein (tapasin), tandem duplicate 2 |
| chr6_-_51811058 | 1.20 |
ENSDART00000031597
|
psmf1
|
proteasome inhibitor subunit 1 |
| chr22_-_36718745 | 1.18 |
|
|
|
| chr15_-_1519615 | 1.17 |
ENSDART00000129356
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr9_+_23954607 | 1.17 |
ENSDART00000145120
|
ENSDARG00000040445
|
ENSDARG00000040445 |
| chr12_-_3018931 | 1.16 |
ENSDART00000139721
|
dcxr
|
dicarbonyl/L-xylulose reductase |
| chr7_+_48025010 | 1.16 |
ENSDART00000052123
|
slc25a44b
|
solute carrier family 25, member 44 b |
| chr20_+_27040570 | 1.15 |
ENSDART00000138369
|
cdca4
|
cell division cycle associated 4 |
| chr25_+_21417388 | 1.13 |
|
|
|
| chr19_-_30265997 | 1.13 |
ENSDART00000024292
|
txlna
|
taxilin alpha |
| chr8_+_13757024 | 1.11 |
|
|
|
| chr22_-_7688508 | 1.09 |
|
|
|
| chr12_-_4563094 | 1.07 |
ENSDART00000152489
|
irf3
|
interferon regulatory factor 3 |
| chr10_+_35208897 | 1.07 |
ENSDART00000063418
|
nsun5
|
NOP2/Sun domain family, member 5 |
| KN150200v1_-_6975 | 1.05 |
|
|
|
| chr24_+_32295048 | 1.04 |
ENSDART00000143570
|
CU207275.1
|
ENSDARG00000092965 |
| chr15_+_43682994 | 1.04 |
ENSDART00000168589
|
ctsc
|
cathepsin C |
| chr16_-_17793032 | 1.04 |
ENSDART00000108581
|
si:dkey-17m8.1
|
si:dkey-17m8.1 |
| chr8_-_11164322 | 1.04 |
ENSDART00000147817
|
fam208b
|
family with sequence similarity 208, member B |
| chr23_-_29431197 | 1.04 |
ENSDART00000156512
|
si:ch211-129o18.4
|
si:ch211-129o18.4 |
| chr9_+_7829443 | 1.03 |
|
|
|
| chr23_-_36350571 | 1.02 |
ENSDART00000110478
|
zgc:174906
|
zgc:174906 |
| chr13_-_26668719 | 1.02 |
ENSDART00000139264
|
vrk2
|
vaccinia related kinase 2 |
| chr18_-_38289008 | 1.01 |
ENSDART00000139519
|
caprin1b
|
cell cycle associated protein 1b |
| chr2_+_42077559 | 1.00 |
ENSDART00000023208
|
ENSDARG00000014209
|
ENSDARG00000014209 |
| chr8_-_11164125 | 1.00 |
ENSDART00000147817
|
fam208b
|
family with sequence similarity 208, member B |
| chr22_-_10540917 | 0.99 |
ENSDART00000140736
|
si:dkey-42i9.6
|
si:dkey-42i9.6 |
| chr24_-_34794463 | 0.98 |
ENSDART00000171009
ENSDART00000170046 |
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr3_+_40022244 | 0.97 |
ENSDART00000102526
|
gfer
|
growth factor, augmenter of liver regeneration (ERV1 homolog, S. cerevisiae) |
| chr17_+_23709676 | 0.97 |
ENSDART00000179026
|
zgc:91976
|
zgc:91976 |
| chr21_-_36710989 | 0.96 |
ENSDART00000086060
|
mrpl22
|
mitochondrial ribosomal protein L22 |
| chr15_-_24653336 | 0.95 |
ENSDART00000123167
|
gemin4
|
gem (nuclear organelle) associated protein 4 |
| chr18_+_13280519 | 0.94 |
ENSDART00000080709
|
plcg2
|
phospholipase C, gamma 2 |
| chr23_+_29431388 | 0.92 |
ENSDART00000027255
|
tardbpl
|
TAR DNA binding protein, like |
| chr2_+_16917277 | 0.90 |
ENSDART00000134135
|
si:dkeyp-13a3.10
|
si:dkeyp-13a3.10 |
| chr14_-_16504730 | 0.90 |
ENSDART00000158396
|
tcirg1b
|
T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3b |
| chr8_+_13757180 | 0.88 |
|
|
|
| chr7_-_667670 | 0.88 |
ENSDART00000159359
|
rela
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr2_-_55584106 | 0.86 |
ENSDART00000169382
ENSDART00000097874 |
tpm4b
|
tropomyosin 4b |
| chr12_+_26970745 | 0.86 |
ENSDART00000153426
|
srcap
|
Snf2-related CREBBP activator protein |
| chr3_-_29557642 | 0.86 |
ENSDART00000151679
|
si:ch73-233k15.2
|
si:ch73-233k15.2 |
| chr5_-_57053661 | 0.85 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr6_+_32849464 | 0.84 |
ENSDART00000157353
|
cyldl
|
cylindromatosis (turban tumor syndrome), like |
| chr18_+_13280623 | 0.84 |
ENSDART00000080709
|
plcg2
|
phospholipase C, gamma 2 |
| chr19_+_4939999 | 0.84 |
ENSDART00000093402
|
cdk12
|
cyclin-dependent kinase 12 |
| chr25_+_31979756 | 0.83 |
ENSDART00000012600
|
galk2
|
galactokinase 2 |
| chr22_-_37630428 | 0.83 |
ENSDART00000149482
|
fxr1
|
fragile X mental retardation, autosomal homolog 1 |
| chr7_-_73628049 | 0.81 |
ENSDART00000123136
|
FP236812.4
|
Histone H2B 1/2 |
| chr18_+_22285328 | 0.81 |
|
|
|
| chr6_-_51810856 | 0.80 |
ENSDART00000031597
|
psmf1
|
proteasome inhibitor subunit 1 |
| chr15_-_5636514 | 0.79 |
ENSDART00000114410
|
wdr62
|
WD repeat domain 62 |
| chr19_+_27864350 | 0.79 |
ENSDART00000052352
|
ppp1r11
|
protein phosphatase 1, regulatory (inhibitor) subunit 11 |
| chr2_+_52529869 | 0.78 |
ENSDART00000111266
|
theg
|
theg spermatid protein |
| chr22_-_6723168 | 0.78 |
|
|
|
| chr19_-_3947553 | 0.78 |
ENSDART00000172271
|
thrap3b
|
thyroid hormone receptor associated protein 3b |
| chr12_-_19029755 | 0.77 |
ENSDART00000057124
|
tefa
|
thyrotrophic embryonic factor a |
| chr18_-_17735227 | 0.77 |
|
|
|
| chr4_-_1908065 | 0.77 |
ENSDART00000144074
|
tm7sf3
|
transmembrane 7 superfamily member 3 |
| chr22_-_7887401 | 0.74 |
ENSDART00000097198
|
sc:d217
|
sc:d217 |
| chr22_-_6723094 | 0.74 |
|
|
|
| chr14_+_21457220 | 0.73 |
ENSDART00000111839
|
kdm2ab
|
lysine (K)-specific demethylase 2Ab |
| chr5_-_37220725 | 0.73 |
ENSDART00000165513
|
AL935065.1
|
ENSDARG00000099693 |
| chr8_-_11164610 | 0.73 |
ENSDART00000147817
|
fam208b
|
family with sequence similarity 208, member B |
| chr24_+_12801404 | 0.72 |
ENSDART00000005105
|
psme1
|
proteasome activator subunit 1 |
| chr25_-_19476079 | 0.71 |
ENSDART00000156811
|
si:ch211-59o9.10
|
si:ch211-59o9.10 |
| chr12_-_26468261 | 0.71 |
ENSDART00000039510
|
tap2t
|
si:dkey-57h18.2 |
| chr15_+_20416637 | 0.66 |
ENSDART00000161047
|
il15l
|
interleukin 15, like |
| chr5_+_43772045 | 0.66 |
ENSDART00000144706
|
BX950187.1
|
ENSDARG00000095282 |
| chr22_-_8075916 | 0.66 |
|
|
|
| chr9_+_56868126 | 0.64 |
ENSDART00000160107
|
CU639413.1
|
ENSDARG00000100275 |
| chr19_-_5946770 | 0.64 |
|
|
|
| chr9_-_19084609 | 0.64 |
ENSDART00000131957
|
crfb2
|
cytokine receptor family member b2 |
| chr3_+_52746019 | 0.64 |
|
|
|
| chr6_-_32849361 | 0.63 |
ENSDART00000129803
|
zc3h3
|
zinc finger CCCH-type containing 3 |
| chr8_+_47197712 | 0.62 |
ENSDART00000137319
|
si:dkeyp-100a1.6
|
si:dkeyp-100a1.6 |
| chr23_-_35297841 | 0.62 |
ENSDART00000145597
|
cmtr1
|
cap methyltransferase 1 |
| chr5_+_32215942 | 0.62 |
ENSDART00000047377
|
crata
|
carnitine O-acetyltransferase a |
| chr12_+_14111340 | 0.61 |
ENSDART00000123741
|
kbtbd2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr21_-_30957754 | 0.60 |
ENSDART00000065503
|
pgap2
|
post-GPI attachment to proteins 2 |
| chr1_+_31608918 | 0.60 |
ENSDART00000137156
|
pudp
|
pseudouridine 5'-phosphatase |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.4 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.8 | 4.8 | GO:0036230 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.6 | 3.2 | GO:0033262 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.5 | 1.5 | GO:0043903 | regulation of symbiosis, encompassing mutualism through parasitism(GO:0043903) interaction with host(GO:0051701) |
| 0.5 | 2.9 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.5 | 1.8 | GO:2000737 | negative regulation of stem cell differentiation(GO:2000737) |
| 0.4 | 1.8 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.4 | 3.2 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.3 | 1.6 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.3 | 2.9 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.3 | 2.0 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.2 | 1.3 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.2 | 1.3 | GO:0016441 | posttranscriptional gene silencing(GO:0016441) posttranscriptional gene silencing by RNA(GO:0035194) |
| 0.2 | 1.4 | GO:0000305 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.2 | 3.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.2 | 1.7 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.2 | 1.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 0.8 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.2 | 1.4 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.2 | 0.6 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 3.4 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 1.1 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 3.3 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.1 | 1.3 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.1 | 0.5 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 0.3 | GO:0032475 | otolith formation(GO:0032475) |
| 0.1 | 0.6 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 0.5 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.1 | 2.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 3.4 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 2.4 | GO:0071482 | cellular response to light stimulus(GO:0071482) |
| 0.1 | 0.7 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.9 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 7.6 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.1 | 1.6 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 1.3 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.1 | 0.5 | GO:0099645 | protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 0.1 | 1.2 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.1 | 0.2 | GO:0001715 | ectodermal cell fate commitment(GO:0001712) ectodermal cell fate specification(GO:0001715) ectodermal cell differentiation(GO:0010668) regulation of ectodermal cell fate specification(GO:0042665) regulation of ectoderm development(GO:2000383) |
| 0.1 | 2.0 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 1.7 | GO:0007632 | visual behavior(GO:0007632) |
| 0.1 | 1.8 | GO:0009225 | nucleotide-sugar metabolic process(GO:0009225) |
| 0.1 | 1.0 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 0.3 | GO:0030262 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 1.4 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.8 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.3 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 1.3 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.8 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.7 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 0.7 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 5.9 | GO:0042278 | purine nucleoside metabolic process(GO:0042278) |
| 0.0 | 0.7 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 7.9 | GO:0042981 | regulation of apoptotic process(GO:0042981) |
| 0.0 | 1.0 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 3.6 | GO:0006915 | apoptotic process(GO:0006915) |
| 0.0 | 1.4 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 1.0 | GO:0001894 | tissue homeostasis(GO:0001894) |
| 0.0 | 0.6 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.2 | GO:0031431 | Dbf4-dependent protein kinase complex(GO:0031431) |
| 0.6 | 1.7 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.4 | 1.2 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.4 | 2.6 | GO:0032797 | SMN complex(GO:0032797) |
| 0.3 | 0.8 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.2 | 0.7 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.2 | 6.0 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 1.6 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 2.8 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 1.3 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 0.9 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.8 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 1.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 2.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.5 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 1.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 2.9 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.0 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 3.6 | GO:0005774 | vacuolar membrane(GO:0005774) |
| 0.0 | 3.0 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 3.2 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 9.0 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 0.4 | GO:0005657 | replication fork(GO:0005657) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 8.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.6 | 3.2 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.6 | 15.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.4 | 1.8 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.4 | 1.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.3 | 2.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.3 | 1.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.3 | 1.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.3 | 1.5 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.3 | 2.9 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.3 | 1.0 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 3.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.2 | 0.6 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.2 | 0.6 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 1.0 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.2 | 3.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 1.5 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.7 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.4 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.1 | 3.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 1.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.7 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.9 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 1.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.3 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.1 | 1.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.7 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 0.9 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 1.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 1.3 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 1.3 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 0.8 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.6 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.1 | 4.8 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.1 | 1.7 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.5 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 2.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.5 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 1.8 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.8 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 1.0 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 1.5 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 1.0 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.7 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.7 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 1.3 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 3.2 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.2 | 3.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 3.3 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 1.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 1.4 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.1 | 1.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 3.0 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.9 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.4 | 1.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 3.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 3.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 3.6 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 2.6 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.1 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |