Project
DANIO-CODE
Navigation
Downloads
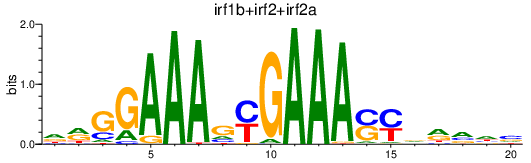
Results for irf1b+irf2+irf2a
Z-value: 2.53
Transcription factors associated with irf1b+irf2+irf2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
irf2a
|
ENSDARG00000007387 | interferon regulatory factor 2a |
|
irf2
|
ENSDARG00000040465 | interferon regulatory factor 2 |
|
irf1b
|
ENSDARG00000043249 | interferon regulatory factor 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| irf2 | dr10_dc_chr14_-_4014025_4014033 | -0.87 | 9.5e-06 | Click! |
| irf2a | dr10_dc_chr1_+_39141680_39141746 | 0.84 | 4.7e-05 | Click! |
| txndc15 | dr10_dc_chr21_-_45800602_45800732 | -0.83 | 6.1e-05 | Click! |
Activity profile of irf1b+irf2+irf2a motif
Sorted Z-values of irf1b+irf2+irf2a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of irf1b+irf2+irf2a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_43868027 | 12.56 |
ENSDART00000112598
ENSDART00000169576 |
otud4
|
OTU deubiquitinase 4 |
| chr1_-_21961282 | 8.42 |
ENSDART00000171830
|
uchl1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr18_-_38289370 | 7.68 |
ENSDART00000139519
|
caprin1b
|
cell cycle associated protein 1b |
| chr14_-_46980330 | 7.58 |
ENSDART00000043751
ENSDART00000141357 |
macrod1
|
MACRO domain containing 1 |
| chr11_+_1558144 | 7.09 |
ENSDART00000154583
|
si:dkey-40c23.2
|
si:dkey-40c23.2 |
| chr1_-_44892852 | 6.11 |
ENSDART00000160961
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr6_+_38898605 | 5.94 |
ENSDART00000029930
|
slc48a1b
|
solute carrier family 48 (heme transporter), member 1b |
| chr11_+_37371675 | 5.83 |
ENSDART00000140502
|
sh2d5
|
SH2 domain containing 5 |
| chr20_-_20922072 | 5.74 |
ENSDART00000142618
ENSDART00000145601 |
ckbb
|
creatine kinase, brain b |
| chr1_-_44892600 | 5.58 |
ENSDART00000149155
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr24_+_32295105 | 5.51 |
ENSDART00000143570
|
CU207275.1
|
ENSDARG00000092965 |
| chr3_+_32285237 | 5.42 |
ENSDART00000157324
|
prrg2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr1_-_55072271 | 5.31 |
ENSDART00000142244
|
dnajb1b
|
DnaJ (Hsp40) homolog, subfamily B, member 1b |
| chr13_-_37504883 | 5.16 |
ENSDART00000108855
|
si:dkey-188i13.7
|
si:dkey-188i13.7 |
| chr8_-_51380691 | 5.13 |
ENSDART00000060628
|
chmp7
|
charged multivesicular body protein 7 |
| chr17_-_18777398 | 5.06 |
ENSDART00000045991
|
vrk1
|
vaccinia related kinase 1 |
| chr5_-_57053687 | 4.99 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr18_-_12889296 | 4.76 |
ENSDART00000130343
|
parp12a
|
poly (ADP-ribose) polymerase family, member 12a |
| chr7_-_24724911 | 4.73 |
ENSDART00000173791
|
rcor2
|
REST corepressor 2 |
| chr4_+_16197 | 4.68 |
ENSDART00000166174
|
ENSDARG00000100660
|
ENSDARG00000100660 |
| chr10_-_45383207 | 4.66 |
ENSDART00000169845
|
pargl
|
poly (ADP-ribose) glycohydrolase, like |
| chr5_-_29782745 | 4.63 |
ENSDART00000125381
|
gig2o
|
grass carp reovirus (GCRV)-induced gene 2o |
| chr15_+_25554119 | 4.57 |
ENSDART00000162482
|
zgc:152863
|
zgc:152863 |
| chr13_-_6123969 | 4.54 |
ENSDART00000161163
|
tuba4l
|
tubulin, alpha 4 like |
| chr5_+_36677193 | 4.45 |
ENSDART00000159402
|
si:dkey-17o15.2
|
si:dkey-17o15.2 |
| chr15_+_20054306 | 4.33 |
ENSDART00000155199
|
zgc:112083
|
zgc:112083 |
| chr3_+_51430252 | 4.29 |
ENSDART00000159493
|
baiap2a
|
BAI1-associated protein 2a |
| chr12_+_17032829 | 4.21 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr10_-_35209022 | 4.18 |
ENSDART00000063434
|
ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr16_-_9911999 | 3.93 |
ENSDART00000154217
|
tapbpl
|
TAP binding protein (tapasin)-like |
| chr18_-_12889369 | 3.92 |
ENSDART00000130343
|
parp12a
|
poly (ADP-ribose) polymerase family, member 12a |
| chr1_-_51567135 | 3.90 |
ENSDART00000143805
|
rab3da
|
RAB3D, member RAS oncogene family, a |
| chr19_+_4135556 | 3.85 |
ENSDART00000162461
|
btr26
|
bloodthirsty-related gene family, member 26 |
| chr15_-_18273640 | 3.84 |
ENSDART00000141508
|
btr16
|
bloodthirsty-related gene family, member 16 |
| chr5_+_21902777 | 3.83 |
ENSDART00000141385
|
si:dkey-27p18.3
|
si:dkey-27p18.3 |
| chr1_-_54505313 | 3.79 |
ENSDART00000100619
ENSDART00000163796 |
LUC7L2
|
zgc:158803 |
| chr24_-_31967674 | 3.74 |
ENSDART00000156060
|
si:ch73-78o10.1
|
si:ch73-78o10.1 |
| chr21_+_31216418 | 3.73 |
ENSDART00000178521
|
asl
|
argininosuccinate lyase |
| chr10_-_210594 | 3.71 |
ENSDART00000059476
|
psmg1
|
proteasome (prosome, macropain) assembly chaperone 1 |
| chr12_+_17032997 | 3.71 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr2_+_36623514 | 3.65 |
ENSDART00000098415
|
ing5b
|
inhibitor of growth family, member 5b |
| chr11_+_36088518 | 3.61 |
|
|
|
| chr8_-_14042703 | 3.59 |
ENSDART00000042867
|
dedd
|
death effector domain containing |
| chr23_+_32015527 | 3.59 |
ENSDART00000088607
|
nemp1
|
nuclear envelope integral membrane protein 1 |
| chr24_+_32295150 | 3.57 |
ENSDART00000143570
|
CU207275.1
|
ENSDARG00000092965 |
| chr22_-_7431710 | 3.54 |
ENSDART00000170630
|
BX511034.7
|
ENSDARG00000104176 |
| chr22_+_31073990 | 3.47 |
ENSDART00000111561
|
zmp:0000000735
|
zmp:0000000735 |
| chr14_+_9175399 | 3.47 |
ENSDART00000054690
ENSDART00000135449 |
st3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr3_+_12087549 | 3.45 |
|
|
|
| chr5_+_36168475 | 3.45 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr16_-_40508858 | 3.44 |
ENSDART00000032389
|
plekhf2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
| chr22_-_7780120 | 3.35 |
ENSDART00000097201
|
si:ch73-44m9.1
|
si:ch73-44m9.1 |
| chr22_-_7412566 | 3.35 |
ENSDART00000161615
ENSDART00000158210 |
BX511034.5
|
ENSDARG00000099295 |
| chr10_-_7862983 | 3.35 |
ENSDART00000160606
ENSDART00000112201 |
mat2aa
|
methionine adenosyltransferase II, alpha a |
| chr2_+_10040130 | 3.32 |
ENSDART00000139639
|
anxa13l
|
annexin A13, like |
| chr19_-_35905513 | 3.28 |
ENSDART00000177052
|
ptp4a2b
|
protein tyrosine phosphatase type IVA, member 2b |
| chr22_+_30041252 | 3.24 |
ENSDART00000165313
|
si:dkey-286j15.3
|
si:dkey-286j15.3 |
| chr13_-_49886891 | 3.21 |
ENSDART00000074230
|
pkz
|
protein kinase containing Z-DNA binding domains |
| chr22_-_7976467 | 3.21 |
ENSDART00000162028
|
sc:d217
|
sc:d217 |
| chr21_-_31215463 | 3.19 |
ENSDART00000170507
|
crcp
|
calcitonin gene-related peptide-receptor component protein |
| chr9_+_30815655 | 3.16 |
ENSDART00000147813
|
tbc1d4
|
TBC1 domain family, member 4 |
| chr6_-_1613740 | 3.00 |
ENSDART00000156305
|
trim107
|
tripartite motif containing 107 |
| chr2_+_35621136 | 2.99 |
ENSDART00000133018
ENSDART00000147278 |
plk3
|
polo-like kinase 3 (Drosophila) |
| chr3_-_21006698 | 2.97 |
ENSDART00000104051
|
cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr7_+_55217087 | 2.95 |
ENSDART00000149915
|
cdt1
|
chromatin licensing and DNA replication factor 1 |
| chr15_+_20416591 | 2.92 |
ENSDART00000161047
|
il15l
|
interleukin 15, like |
| chr5_-_29782945 | 2.85 |
ENSDART00000125381
|
gig2o
|
grass carp reovirus (GCRV)-induced gene 2o |
| chr18_-_23847383 | 2.84 |
ENSDART00000155068
|
BX936364.1
|
ENSDARG00000097764 |
| KN150200v1_-_6975 | 2.77 |
|
|
|
| chr5_+_57056091 | 2.75 |
ENSDART00000134684
|
fdxacb1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr7_+_66660882 | 2.73 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr1_-_54505112 | 2.72 |
ENSDART00000100619
ENSDART00000163796 |
LUC7L2
|
zgc:158803 |
| chr6_+_36862078 | 2.68 |
ENSDART00000104160
|
ENSDARG00000070746
|
ENSDARG00000070746 |
| chr13_+_2345595 | 2.68 |
|
|
|
| chr5_-_29782972 | 2.66 |
ENSDART00000125381
|
gig2o
|
grass carp reovirus (GCRV)-induced gene 2o |
| chr9_-_23954640 | 2.65 |
ENSDART00000027212
|
ENSDARG00000006848
|
ENSDARG00000006848 |
| chr15_+_20416518 | 2.64 |
ENSDART00000161047
|
il15l
|
interleukin 15, like |
| chr12_-_4800362 | 2.64 |
ENSDART00000172093
|
si:ch211-93e11.8
|
si:ch211-93e11.8 |
| chr2_-_21512154 | 2.63 |
ENSDART00000135417
|
bmi1b
|
bmi1 polycomb ring finger oncogene 1b |
| chr23_+_33792145 | 2.63 |
ENSDART00000024695
|
dazap2
|
DAZ associated protein 2 |
| chr11_-_44734403 | 2.61 |
ENSDART00000160656
|
afmid
|
arylformamidase |
| chr3_-_39117855 | 2.56 |
ENSDART00000102678
|
nmt1a
|
N-myristoyltransferase 1a |
| chr9_+_22015827 | 2.55 |
ENSDART00000101985
|
zgc:162944
|
zgc:162944 |
| chr5_+_27697056 | 2.54 |
ENSDART00000132070
|
vamp5
|
vesicle-associated membrane protein 5 |
| chr1_-_44892558 | 2.53 |
ENSDART00000149155
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr22_-_8144599 | 2.52 |
ENSDART00000156571
|
si:ch73-44m9.3
|
si:ch73-44m9.3 |
| chr15_-_1519615 | 2.52 |
ENSDART00000129356
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr13_+_11418060 | 2.51 |
ENSDART00000166908
|
desi2
|
desumoylating isopeptidase 2 |
| chr6_+_49054117 | 2.50 |
ENSDART00000011876
|
sycp1
|
synaptonemal complex protein 1 |
| chr15_-_29229170 | 2.49 |
ENSDART00000138449
|
xaf1
|
XIAP associated factor 1 |
| chr19_+_38580804 | 2.49 |
ENSDART00000160756
|
phf14
|
PHD finger protein 14 |
| chr17_-_42523585 | 2.47 |
ENSDART00000154755
|
prkd3
|
protein kinase D3 |
| chr24_+_16838877 | 2.47 |
ENSDART00000145520
|
eif2s3
|
eukaryotic translation initiation factor 2, subunit 3 gamma |
| chr5_-_23092592 | 2.46 |
ENSDART00000024815
|
fam76b
|
family with sequence similarity 76, member B |
| chr2_-_26820425 | 2.45 |
ENSDART00000132125
|
acadm
|
acyl-CoA dehydrogenase, C-4 to C-12 straight chain |
| chr12_-_37274632 | 2.45 |
ENSDART00000152951
|
cdc42ep4b
|
CDC42 effector protein (Rho GTPase binding) 4b |
| chr10_-_25366450 | 2.44 |
ENSDART00000123820
|
tmem135
|
transmembrane protein 135 |
| chr14_+_36074512 | 2.44 |
ENSDART00000015761
|
neil3
|
nei-like DNA glycosylase 3 |
| chr3_-_19890717 | 2.44 |
ENSDART00000104118
ENSDART00000170199 |
atxn7l3
|
ataxin 7-like 3 |
| chr3_+_32530476 | 2.42 |
ENSDART00000171556
|
si:dkey-16l2.16
|
si:dkey-16l2.16 |
| chr10_+_35209025 | 2.42 |
ENSDART00000126105
|
nsun5
|
NOP2/Sun domain family, member 5 |
| chr13_+_7055611 | 2.39 |
ENSDART00000148240
|
exoc3l4
|
exocyst complex component 3-like 4 |
| chr12_+_1247340 | 2.37 |
|
|
|
| chr11_-_36088336 | 2.37 |
ENSDART00000141477
|
psma5
|
proteasome subunit alpha 5 |
| chr18_-_23847340 | 2.37 |
ENSDART00000155068
|
BX936364.1
|
ENSDARG00000097764 |
| chr7_-_69883542 | 2.36 |
|
|
|
| KN150335v1_-_25856 | 2.31 |
ENSDART00000164352
|
zgc:171497
|
zgc:171497 |
| chr12_-_26468261 | 2.31 |
ENSDART00000039510
|
tap2t
|
si:dkey-57h18.2 |
| chr12_+_17032963 | 2.30 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr17_-_25612341 | 2.30 |
ENSDART00000126201
ENSDART00000105503 |
rab3gap2
|
RAB3 GTPase activating protein subunit 2 (non-catalytic) |
| chr6_+_12810986 | 2.29 |
ENSDART00000104757
ENSDART00000165896 |
casp8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr15_+_14656645 | 2.28 |
ENSDART00000162350
|
FBXO46
|
F-box protein 46 |
| chr4_-_26106485 | 2.26 |
ENSDART00000100611
|
si:ch211-244b2.3
|
si:ch211-244b2.3 |
| chr9_+_8919663 | 2.26 |
ENSDART00000134954
|
carkd
|
carbohydrate kinase domain containing |
| chr9_-_41151768 | 2.23 |
ENSDART00000135805
|
BX323586.2
|
ENSDARG00000092912 |
| chr23_+_38228263 | 2.22 |
ENSDART00000137969
|
zgc:112994
|
zgc:112994 |
| chr21_+_20286289 | 2.21 |
ENSDART00000153992
ENSDART00000156867 |
AL844141.1
|
ENSDARG00000096964 |
| chr15_+_17162691 | 2.20 |
|
|
|
| chr13_+_26573404 | 2.20 |
ENSDART00000142483
|
fancl
|
Fanconi anemia, complementation group L |
| chr24_-_34794538 | 2.20 |
ENSDART00000171009
ENSDART00000170046 |
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr7_-_69883581 | 2.17 |
|
|
|
| chr24_+_32295048 | 2.17 |
ENSDART00000143570
|
CU207275.1
|
ENSDARG00000092965 |
| chr6_-_1613915 | 2.16 |
ENSDART00000112118
|
trim107
|
tripartite motif containing 107 |
| chr5_+_57056176 | 2.16 |
ENSDART00000134684
|
fdxacb1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr25_-_25455861 | 2.15 |
ENSDART00000150412
|
irf7
|
interferon regulatory factor 7 |
| chr20_+_29328615 | 2.15 |
ENSDART00000147064
|
aven
|
apoptosis, caspase activation inhibitor |
| chr6_-_54425831 | 2.12 |
ENSDART00000017230
|
snrpc
|
small nuclear ribonucleoprotein polypeptide C |
| chr16_-_9562744 | 2.11 |
ENSDART00000126154
|
prpf3
|
PRP3 pre-mRNA processing factor 3 homolog (yeast) |
| chr7_-_71343686 | 2.11 |
ENSDART00000128678
|
mettl4
|
methyltransferase like 4 |
| chr15_-_29229135 | 2.10 |
ENSDART00000138449
|
xaf1
|
XIAP associated factor 1 |
| chr16_+_14119938 | 2.09 |
ENSDART00000059928
|
fdps
|
farnesyl diphosphate synthase (farnesyl pyrophosphate synthetase, dimethylallyltranstransferase, geranyltranstransferase) |
| chr5_-_57053661 | 2.09 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr22_-_34639168 | 2.09 |
|
|
|
| chr8_-_21020134 | 2.09 |
ENSDART00000136561
|
si:dkeyp-82a1.6
|
si:dkeyp-82a1.6 |
| chr12_-_4563094 | 2.08 |
ENSDART00000152489
|
irf3
|
interferon regulatory factor 3 |
| chr9_+_29806055 | 2.08 |
ENSDART00000143493
|
phf11
|
PHD finger protein 11 |
| chr1_-_51567283 | 2.06 |
ENSDART00000023757
|
rab3da
|
RAB3D, member RAS oncogene family, a |
| chr17_-_42523635 | 2.06 |
ENSDART00000154755
|
prkd3
|
protein kinase D3 |
| chr9_-_40881885 | 2.03 |
ENSDART00000141979
|
bard1
|
BRCA1 associated RING domain 1 |
| chr5_+_40235387 | 2.03 |
ENSDART00000147767
|
si:dkey-3h3.3
|
si:dkey-3h3.3 |
| chr24_-_13205000 | 2.02 |
ENSDART00000134482
|
terf1
|
telomeric repeat binding factor (NIMA-interacting) 1 |
| chr4_-_19911139 | 2.02 |
|
|
|
| chr6_-_1613994 | 2.02 |
ENSDART00000112118
|
trim107
|
tripartite motif containing 107 |
| chr25_-_19476079 | 2.01 |
ENSDART00000156811
|
si:ch211-59o9.10
|
si:ch211-59o9.10 |
| chr18_-_38289008 | 2.01 |
ENSDART00000139519
|
caprin1b
|
cell cycle associated protein 1b |
| chr13_+_11417882 | 2.00 |
ENSDART00000034935
|
desi2
|
desumoylating isopeptidase 2 |
| chr11_+_39664664 | 2.00 |
ENSDART00000102734
ENSDART00000137516 |
vamp3
|
vesicle-associated membrane protein 3 (cellubrevin) |
| chr22_-_7887341 | 1.99 |
ENSDART00000097198
|
sc:d217
|
sc:d217 |
| chr23_-_36350571 | 1.98 |
ENSDART00000110478
|
zgc:174906
|
zgc:174906 |
| chr20_+_44684340 | 1.98 |
ENSDART00000149775
|
atad2b
|
ATPase family, AAA domain containing 2B |
| chr16_-_31834774 | 1.97 |
|
|
|
| chr7_+_19348229 | 1.97 |
ENSDART00000007310
ENSDART00000166355 |
zgc:171731
|
zgc:171731 |
| chr14_+_31278584 | 1.96 |
ENSDART00000158875
ENSDART00000026195 |
slc9a6a
|
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6a |
| chr25_-_27222696 | 1.95 |
ENSDART00000123590
|
si:ch211-91p5.3
|
si:ch211-91p5.3 |
| chr10_+_21487218 | 1.93 |
ENSDART00000147215
ENSDART00000019252 |
fbxw11b
|
F-box and WD repeat domain containing 11b |
| chr5_+_69007907 | 1.92 |
ENSDART00000154507
|
si:ch211-275j6.5
|
si:ch211-275j6.5 |
| chr13_-_40592360 | 1.91 |
|
|
|
| chr5_-_56953716 | 1.91 |
ENSDART00000167892
|
zgc:193711
|
zgc:193711 |
| chr7_-_40307611 | 1.90 |
ENSDART00000173926
ENSDART00000010035 |
dnajb6b
|
DnaJ (Hsp40) homolog, subfamily B, member 6b |
| chr23_-_36350288 | 1.90 |
ENSDART00000110478
|
zgc:174906
|
zgc:174906 |
| chr18_-_14910429 | 1.90 |
ENSDART00000099701
|
selo
|
selenoprotein O |
| chr3_+_1054368 | 1.88 |
ENSDART00000153693
|
si:ch73-166o21.1
|
si:ch73-166o21.1 |
| chr5_-_56953587 | 1.88 |
ENSDART00000167892
|
zgc:193711
|
zgc:193711 |
| chr5_+_44204845 | 1.87 |
ENSDART00000141198
|
ctsla
|
cathepsin La |
| chr18_+_14715573 | 1.87 |
ENSDART00000108469
|
spata2l
|
spermatogenesis associated 2-like |
| chr15_-_16447990 | 1.86 |
ENSDART00000154504
|
fam222bb
|
family with sequence similarity 222, member Bb |
| chr14_+_11864388 | 1.86 |
ENSDART00000146521
|
rhogd
|
ras homolog gene family, member Gd |
| chr2_-_8813671 | 1.86 |
ENSDART00000138223
|
si:ch211-71m22.1
|
si:ch211-71m22.1 |
| chr9_-_35824470 | 1.85 |
ENSDART00000140356
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr5_+_32215942 | 1.84 |
ENSDART00000047377
|
crata
|
carnitine O-acetyltransferase a |
| chr10_-_21971084 | 1.84 |
ENSDART00000174954
|
CABZ01031892.1
|
ENSDARG00000107280 |
| chr5_+_45295180 | 1.84 |
ENSDART00000083937
|
polk
|
polymerase (DNA directed) kappa |
| chr11_-_40239946 | 1.83 |
ENSDART00000165394
|
si:dkeyp-61b2.1
|
si:dkeyp-61b2.1 |
| chr10_-_7862767 | 1.83 |
ENSDART00000059021
|
mat2aa
|
methionine adenosyltransferase II, alpha a |
| chr21_+_19283166 | 1.76 |
ENSDART00000063621
|
abraxas2a
|
abraxas 2a, BRISC complex subunit |
| chr8_-_51380846 | 1.76 |
ENSDART00000060628
|
chmp7
|
charged multivesicular body protein 7 |
| chr1_-_529071 | 1.75 |
ENSDART00000147610
|
si:ch73-41e3.7
|
si:ch73-41e3.7 |
| chr22_+_2049719 | 1.74 |
|
|
|
| chr14_+_11863984 | 1.74 |
ENSDART00000129953
|
rhogd
|
ras homolog gene family, member Gd |
| chr16_+_17762256 | 1.73 |
ENSDART00000128672
|
tmem238b
|
transmembrane protein 238b |
| chr23_-_45011264 | 1.72 |
ENSDART00000129138
|
mogat3a
|
monoacylglycerol O-acyltransferase 3a |
| chr5_-_5848617 | 1.71 |
|
|
|
| chr18_+_13280400 | 1.70 |
ENSDART00000080709
|
plcg2
|
phospholipase C, gamma 2 |
| chr5_+_44204661 | 1.69 |
ENSDART00000122288
|
ctsla
|
cathepsin La |
| chr21_-_21477462 | 1.69 |
ENSDART00000031205
|
pvrl3b
|
poliovirus receptor-related 3b |
| chr8_+_47108614 | 1.68 |
ENSDART00000142979
|
arhgef16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr13_-_33096999 | 1.68 |
ENSDART00000057386
|
golga5
|
golgin A5 |
| chr16_-_40508938 | 1.67 |
ENSDART00000032389
|
plekhf2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
| chr19_-_34529880 | 1.66 |
ENSDART00000158677
ENSDART00000112004 |
si:dkey-184p18.2
|
si:dkey-184p18.2 |
| chr1_+_22798407 | 1.66 |
ENSDART00000134576
|
si:dkeyp-26a9.2
|
si:dkeyp-26a9.2 |
| chr14_+_11863939 | 1.63 |
ENSDART00000129953
|
rhogd
|
ras homolog gene family, member Gd |
| chr19_+_15581145 | 1.63 |
ENSDART00000079014
|
pdik1l
|
PDLIM1 interacting kinase 1 like |
| chr15_-_14147062 | 1.63 |
ENSDART00000147796
|
trappc6bl
|
trafficking protein particle complex 6b-like |
| chr4_-_12979951 | 1.61 |
ENSDART00000013839
|
tmbim4
|
transmembrane BAX inhibitor motif containing 4 |
| chr13_+_17564330 | 1.59 |
ENSDART00000079778
|
ifit8
|
interferon-induced protein with tetratricopeptide repeats 8 |
| chr9_-_34450885 | 1.59 |
ENSDART00000012385
|
me3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr12_-_20494169 | 1.58 |
ENSDART00000105362
|
snx11
|
sorting nexin 11 |
| chr2_-_21512285 | 1.58 |
ENSDART00000135417
|
bmi1b
|
bmi1 polycomb ring finger oncogene 1b |
| chr2_-_8813612 | 1.57 |
ENSDART00000138223
|
si:ch211-71m22.1
|
si:ch211-71m22.1 |
| chr9_-_23954690 | 1.57 |
ENSDART00000027212
|
ENSDARG00000006848
|
ENSDARG00000006848 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 12.6 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 2.9 | 8.7 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 1.2 | 14.7 | GO:0090308 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 1.1 | 6.7 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 1.0 | 6.2 | GO:0036230 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 1.0 | 1.0 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.9 | 3.6 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.9 | 3.5 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.8 | 2.5 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.8 | 2.3 | GO:0043903 | regulation of symbiosis, encompassing mutualism through parasitism(GO:0043903) interaction with host(GO:0051701) |
| 0.8 | 3.0 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.7 | 5.7 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.7 | 2.9 | GO:1904969 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.7 | 2.1 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.7 | 2.0 | GO:0019376 | galactosylceramide catabolic process(GO:0006683) galactolipid catabolic process(GO:0019376) |
| 0.7 | 7.3 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.6 | 2.6 | GO:0018008 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.6 | 2.5 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.6 | 2.5 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) medium-chain fatty acid catabolic process(GO:0051793) |
| 0.6 | 3.6 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.6 | 8.3 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.5 | 2.2 | GO:2000737 | negative regulation of stem cell differentiation(GO:2000737) |
| 0.5 | 4.3 | GO:2000251 | regulation of actin cytoskeleton reorganization(GO:2000249) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.5 | 2.1 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.5 | 6.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.5 | 7.1 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.5 | 1.8 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.4 | 1.3 | GO:0070292 | N-acylphosphatidylethanolamine metabolic process(GO:0070292) |
| 0.4 | 3.7 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.4 | 2.4 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.4 | 2.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.4 | 5.4 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.4 | 1.5 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.4 | 4.5 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.4 | 1.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.3 | 3.0 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.3 | 1.3 | GO:0043247 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.3 | 5.2 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.3 | 3.5 | GO:0031272 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.3 | 3.1 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.3 | 1.7 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.3 | 3.9 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.3 | 1.3 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.2 | 1.9 | GO:1903573 | negative regulation of response to endoplasmic reticulum stress(GO:1903573) |
| 0.2 | 0.7 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.2 | 0.9 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.2 | 1.8 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.2 | 3.6 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.2 | 1.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 1.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.2 | 3.5 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.2 | 6.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.2 | 2.7 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.2 | 5.1 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.2 | 4.7 | GO:0009225 | nucleotide-sugar metabolic process(GO:0009225) |
| 0.2 | 3.8 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.2 | 4.8 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.1 | 1.9 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.1 | 4.0 | GO:0010833 | telomere maintenance via telomere lengthening(GO:0010833) |
| 0.1 | 13.1 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 0.9 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 3.5 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 1.5 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.1 | 0.9 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 1.0 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.1 | 2.2 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.1 | 2.2 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.1 | 0.9 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.1 | 0.6 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.1 | 0.5 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 0.6 | GO:0019079 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism metabolic process(GO:0044033) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.2 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.1 | 0.9 | GO:0072078 | renal tubule morphogenesis(GO:0061333) nephron tubule morphogenesis(GO:0072078) nephron epithelium morphogenesis(GO:0072088) |
| 0.1 | 9.6 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.1 | 1.0 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 1.7 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.1 | 1.5 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 0.6 | GO:0099633 | protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 0.1 | 1.0 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.1 | 0.9 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 4.7 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 1.1 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.1 | 1.4 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.1 | 0.2 | GO:2000058 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.1 | 0.6 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.1 | 0.4 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.7 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 | 2.4 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 2.4 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.1 | 2.5 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.1 | 0.4 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 4.4 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.3 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.7 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 1.5 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) negative regulation of gene silencing(GO:0060969) |
| 0.0 | 1.4 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 21.1 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.5 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.2 | GO:1903201 | regulation of cellular response to oxidative stress(GO:1900407) negative regulation of cellular response to oxidative stress(GO:1900408) negative regulation of response to oxidative stress(GO:1902883) regulation of oxidative stress-induced cell death(GO:1903201) negative regulation of oxidative stress-induced cell death(GO:1903202) |
| 0.0 | 0.8 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.5 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 2.2 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.8 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.2 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.0 | 0.9 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 1.1 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.4 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 0.8 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 0.2 | GO:0006692 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 0.9 | GO:0007031 | peroxisome organization(GO:0007031) |
| 0.0 | 2.0 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.0 | 0.1 | GO:0071265 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.5 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.1 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.4 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 1.6 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 0.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.3 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 3.5 | GO:0042278 | purine nucleoside metabolic process(GO:0042278) |
| 0.0 | 1.1 | GO:0000819 | sister chromatid segregation(GO:0000819) |
| 0.0 | 1.0 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 | 0.2 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 3.5 | GO:0000278 | mitotic cell cycle(GO:0000278) |
| 0.0 | 4.4 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 0.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 1.0 | GO:0001894 | tissue homeostasis(GO:0001894) |
| 0.0 | 1.2 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.0 | 0.3 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.7 | GO:0071560 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.0 | 0.6 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 0.3 | GO:0099518 | vesicle cytoskeletal trafficking(GO:0099518) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 6.0 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.8 | 2.5 | GO:0000801 | central element(GO:0000801) |
| 0.6 | 7.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.5 | 1.4 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.5 | 6.8 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.4 | 1.3 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.4 | 2.5 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.4 | 2.0 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.3 | 4.0 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.2 | 2.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.2 | 1.9 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 4.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 0.9 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.2 | 5.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.2 | 1.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.2 | 2.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 8.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.2 | 4.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 1.7 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 3.4 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.1 | 4.3 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 1.5 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 0.9 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 3.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 5.8 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 1.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 6.9 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 4.5 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 4.6 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 10.5 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.1 | 5.4 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.2 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.1 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 4.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 1.1 | GO:0030126 | COPI vesicle coat(GO:0030126) COPI-coated vesicle membrane(GO:0030663) |
| 0.1 | 0.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.4 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.3 | GO:0030880 | RNA polymerase complex(GO:0030880) |
| 0.0 | 1.0 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.4 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 3.2 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.4 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 1.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 3.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 7.3 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 3.4 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.5 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 1.4 | GO:0005774 | vacuolar membrane(GO:0005774) |
| 0.0 | 1.8 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 6.6 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 0.3 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.1 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | GO:0070042 | rRNA (uridine-N3-)-methyltransferase activity(GO:0070042) |
| 1.2 | 3.5 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 1.2 | 12.7 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 1.0 | 6.0 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.9 | 3.7 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.9 | 25.9 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.9 | 3.6 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.9 | 3.5 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.8 | 2.4 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.7 | 5.7 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.7 | 2.0 | GO:0004336 | galactosylceramidase activity(GO:0004336) |
| 0.7 | 7.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.6 | 3.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.6 | 2.6 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.6 | 1.8 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.6 | 3.0 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.5 | 1.6 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.4 | 2.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.4 | 3.0 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.4 | 1.3 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.4 | 2.5 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.4 | 1.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.3 | 1.4 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.3 | 4.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.3 | 3.0 | GO:0097200 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.3 | 3.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.3 | 1.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.3 | 1.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.3 | 2.3 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.2 | 2.4 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.2 | 4.5 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.2 | 1.2 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.2 | 1.7 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.2 | 3.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.2 | 1.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.2 | 4.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 6.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 1.4 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.2 | 1.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.2 | 3.8 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.2 | 1.0 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.2 | 1.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.2 | 2.5 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.2 | 3.2 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.2 | 0.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 12.9 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 0.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 1.5 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 0.4 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.1 | 3.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.5 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 4.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 2.9 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 0.9 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 1.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 1.0 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 1.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 2.1 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 1.7 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 3.2 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 0.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.7 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 0.2 | GO:0047522 | 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.1 | 3.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.3 | GO:0046922 | peptide-O-fucosyltransferase activity(GO:0046922) |
| 0.1 | 3.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 1.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 0.7 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 2.5 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 0.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.6 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.1 | 0.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 21.5 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 2.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 6.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.8 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.3 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 4.2 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 3.4 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 1.9 | GO:0005126 | cytokine receptor binding(GO:0005126) |
| 0.0 | 0.3 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 6.1 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 2.3 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.7 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.4 | GO:0070035 | ATP-dependent DNA helicase activity(GO:0004003) ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.9 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 8.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.7 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.4 | GO:0042393 | histone binding(GO:0042393) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.5 | 8.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.3 | 5.9 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.2 | 4.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 3.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 4.2 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 6.6 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 1.9 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 4.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 2.0 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 2.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.2 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 2.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.6 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.6 | 2.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.3 | 7.1 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.3 | 3.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.3 | 2.0 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 4.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 3.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 4.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 3.0 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 1.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 6.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 2.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 2.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.5 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 2.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 0.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 3.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 0.9 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 1.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 0.5 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 0.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 3.0 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 3.0 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |