Project
DANIO-CODE
Navigation
Downloads
Results for irf4a+irf4b
Z-value: 0.41
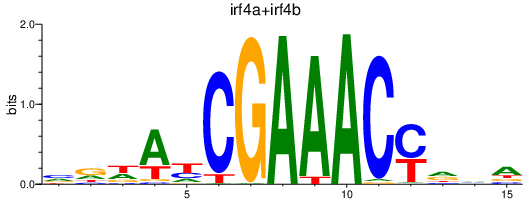
Transcription factors associated with irf4a+irf4b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
irf4a
|
ENSDARG00000006560 | interferon regulatory factor 4a |
|
irf4b
|
ENSDARG00000055374 | interferon regulatory factor 4b |
Activity profile of irf4a+irf4b motif
Sorted Z-values of irf4a+irf4b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of irf4a+irf4b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_23954640 | 0.94 |
ENSDART00000027212
|
ENSDARG00000006848
|
ENSDARG00000006848 |
| chr1_-_54505112 | 0.75 |
ENSDART00000100619
ENSDART00000163796 |
LUC7L2
|
zgc:158803 |
| chr1_-_54505313 | 0.73 |
ENSDART00000100619
ENSDART00000163796 |
LUC7L2
|
zgc:158803 |
| chr25_+_15901398 | 0.66 |
ENSDART00000140047
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr16_+_19831573 | 0.53 |
ENSDART00000135359
|
macc1
|
metastasis associated in colon cancer 1 |
| chr9_-_23954690 | 0.49 |
ENSDART00000027212
|
ENSDARG00000006848
|
ENSDARG00000006848 |
| chr13_+_24132617 | 0.47 |
ENSDART00000135992
|
abcb10
|
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
| chr24_-_23793117 | 0.44 |
|
|
|
| chr16_-_29451902 | 0.44 |
|
|
|
| chr4_-_17680826 | 0.43 |
ENSDART00000066997
|
dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr21_+_17265099 | 0.43 |
ENSDART00000145057
|
tsc1b
|
tuberous sclerosis 1b |
| chr7_-_69576406 | 0.43 |
|
|
|
| chr6_-_10492724 | 0.40 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr21_+_20974216 | 0.39 |
ENSDART00000079692
|
ndr1
|
nodal-related 1 |
| chr22_-_7748347 | 0.39 |
ENSDART00000097276
|
si:ch73-44m9.3
|
si:ch73-44m9.3 |
| chr5_+_27659432 | 0.38 |
ENSDART00000087684
|
ncaph
|
non-SMC condensin I complex, subunit H |
| chr19_+_44073792 | 0.38 |
ENSDART00000051723
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr22_-_7431710 | 0.36 |
ENSDART00000170630
|
BX511034.7
|
ENSDARG00000104176 |
| chr19_-_15516318 | 0.35 |
ENSDART00000151454
|
serinc2
|
serine incorporator 2 |
| chr9_-_23954761 | 0.35 |
ENSDART00000027212
|
ENSDARG00000006848
|
ENSDARG00000006848 |
| chr10_-_33435736 | 0.35 |
ENSDART00000023509
|
ska2
|
spindle and kinetochore associated complex subunit 2 |
| chr9_+_23954669 | 0.35 |
ENSDART00000145120
|
ENSDARG00000040445
|
ENSDARG00000040445 |
| chr18_+_44539099 | 0.34 |
ENSDART00000140672
|
st14a
|
suppression of tumorigenicity 14 (colon carcinoma) a |
| chr7_-_69576628 | 0.34 |
|
|
|
| chr6_-_1613994 | 0.34 |
ENSDART00000112118
|
trim107
|
tripartite motif containing 107 |
| chr9_+_23954807 | 0.33 |
ENSDART00000145120
|
ENSDARG00000040445
|
ENSDARG00000040445 |
| chr7_-_69576270 | 0.33 |
|
|
|
| chr24_-_11326961 | 0.32 |
ENSDART00000082264
|
pxdc1b
|
PX domain containing 1b |
| chr20_+_35156812 | 0.32 |
|
|
|
| chr12_+_26970745 | 0.31 |
ENSDART00000153426
|
srcap
|
Snf2-related CREBBP activator protein |
| chr25_+_16549708 | 0.31 |
ENSDART00000110426
|
cecr2
|
cat eye syndrome chromosome region, candidate 2 |
| chr1_-_36039634 | 0.30 |
ENSDART00000148386
|
prmt9
|
protein arginine methyltransferase 9 |
| chr1_-_54504911 | 0.28 |
ENSDART00000100619
ENSDART00000163796 |
LUC7L2
|
zgc:158803 |
| chr10_-_17593407 | 0.28 |
ENSDART00000145077
|
ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr19_+_44073707 | 0.27 |
ENSDART00000051723
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr18_+_44538877 | 0.27 |
ENSDART00000086952
|
st14a
|
suppression of tumorigenicity 14 (colon carcinoma) a |
| chr6_+_9735186 | 0.27 |
ENSDART00000065471
|
abi2b
|
abl-interactor 2b |
| chr8_-_18193491 | 0.26 |
ENSDART00000143036
|
si:ch211-241d21.5
|
si:ch211-241d21.5 |
| chr11_-_30261689 | 0.26 |
ENSDART00000101667
|
map4k3a
|
mitogen-activated protein kinase kinase kinase kinase 3a |
| chr13_+_17564330 | 0.26 |
ENSDART00000079778
|
ifit8
|
interferon-induced protein with tetratricopeptide repeats 8 |
| chr3_-_26040846 | 0.26 |
ENSDART00000113890
|
rabep2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr10_+_8143021 | 0.26 |
ENSDART00000138875
|
pstpip2
|
proline-serine-threonine phosphatase interacting protein 2 |
| chr25_+_15901561 | 0.25 |
ENSDART00000140047
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr25_+_15901662 | 0.24 |
ENSDART00000140047
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr17_-_22532811 | 0.24 |
ENSDART00000079401
|
si:ch211-125o16.4
|
si:ch211-125o16.4 |
| chr9_+_23954607 | 0.24 |
ENSDART00000145120
|
ENSDARG00000040445
|
ENSDARG00000040445 |
| chr23_-_29627060 | 0.23 |
ENSDART00000166554
|
ube4b
|
ubiquitination factor E4B, UFD2 homolog (S. cerevisiae) |
| chr19_+_791656 | 0.23 |
ENSDART00000138406
|
tmem79a
|
transmembrane protein 79a |
| chr16_+_27449058 | 0.21 |
ENSDART00000132329
|
stx17
|
syntaxin 17 |
| chr6_-_10492439 | 0.21 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr21_-_22507089 | 0.21 |
ENSDART00000177084
|
myo5b
|
myosin VB |
| chr6_-_1613740 | 0.21 |
ENSDART00000156305
|
trim107
|
tripartite motif containing 107 |
| chr25_+_35540123 | 0.21 |
ENSDART00000126326
|
rpgrip1l
|
RPGRIP1-like |
| chr10_+_8142976 | 0.21 |
ENSDART00000123447
|
pstpip2
|
proline-serine-threonine phosphatase interacting protein 2 |
| chr9_+_23192282 | 0.20 |
ENSDART00000021060
|
eaf2
|
ELL associated factor 2 |
| chr15_-_41777233 | 0.19 |
ENSDART00000154230
|
ftr90
|
finTRIM family, member 90 |
| chr4_-_29059042 | 0.19 |
ENSDART00000169971
|
CR387997.1
|
ENSDARG00000099486 |
| chr13_-_24132195 | 0.19 |
|
|
|
| chr18_-_44539130 | 0.19 |
|
|
|
| chr2_+_2976602 | 0.18 |
ENSDART00000081976
|
rock1
|
Rho-associated, coiled-coil containing protein kinase 1 |
| chr6_-_1613915 | 0.18 |
ENSDART00000112118
|
trim107
|
tripartite motif containing 107 |
| chr10_+_110941 | 0.18 |
ENSDART00000135572
|
pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr11_+_6209097 | 0.17 |
ENSDART00000156437
|
BX248501.1
|
ENSDARG00000074519 |
| chr2_+_2976671 | 0.16 |
ENSDART00000081976
|
rock1
|
Rho-associated, coiled-coil containing protein kinase 1 |
| chr6_-_923389 | 0.16 |
ENSDART00000167390
|
zgc:113442
|
zgc:113442 |
| chr16_-_41716834 | 0.15 |
ENSDART00000084528
|
atp2c1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr19_-_30265997 | 0.14 |
ENSDART00000024292
|
txlna
|
taxilin alpha |
| chr10_-_20524586 | 0.13 |
|
|
|
| chr7_-_17129072 | 0.12 |
|
|
|
| chr21_-_40025958 | 0.12 |
ENSDART00000124416
|
med31
|
mediator complex subunit 31 |
| chr10_+_28273581 | 0.12 |
ENSDART00000131003
|
rnft1
|
ring finger protein, transmembrane 1 |
| chr23_-_35297841 | 0.12 |
ENSDART00000145597
|
cmtr1
|
cap methyltransferase 1 |
| chr3_-_26040615 | 0.12 |
ENSDART00000113890
|
rabep2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr14_-_33245562 | 0.11 |
ENSDART00000023540
|
sash3
|
SAM and SH3 domain containing 3 |
| chr8_-_51354135 | 0.10 |
ENSDART00000170619
|
kansl3
|
KAT8 regulatory NSL complex subunit 3 |
| chr2_+_2976973 | 0.10 |
ENSDART00000163587
|
rock1
|
Rho-associated, coiled-coil containing protein kinase 1 |
| chr15_+_34211736 | 0.09 |
ENSDART00000163513
|
arl4aa
|
ADP-ribosylation factor-like 4aa |
| chr7_+_19486714 | 0.08 |
ENSDART00000089333
|
tkfc
|
triokinase/FMN cyclase |
| chr11_-_30261559 | 0.06 |
ENSDART00000101667
|
map4k3a
|
mitogen-activated protein kinase kinase kinase kinase 3a |
| chr13_+_24132494 | 0.05 |
ENSDART00000135992
|
abcb10
|
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
| chr16_+_10666906 | 0.05 |
ENSDART00000091241
|
si:ch73-22o12.1
|
si:ch73-22o12.1 |
| chr19_+_1743359 | 0.05 |
ENSDART00000166744
|
dennd3a
|
DENN/MADD domain containing 3a |
| chr19_+_44073507 | 0.04 |
ENSDART00000139151
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr19_-_30266029 | 0.04 |
ENSDART00000024292
|
txlna
|
taxilin alpha |
| chr22_+_748535 | 0.04 |
ENSDART00000125347
|
cry1bb
|
cryptochrome circadian clock 1bb |
| chr2_+_30506215 | 0.03 |
ENSDART00000115271
|
march6
|
membrane-associated ring finger (C3HC4) 6 |
| chr6_+_15635738 | 0.03 |
ENSDART00000128939
|
iqca1
|
IQ motif containing with AAA domain 1 |
| chr3_-_29557642 | 0.03 |
ENSDART00000151679
|
si:ch73-233k15.2
|
si:ch73-233k15.2 |
| chr21_+_37985536 | 0.02 |
ENSDART00000085728
|
klf8
|
Kruppel-like factor 8 |
| chr4_+_11465367 | 0.02 |
ENSDART00000008584
|
gdi2
|
GDP dissociation inhibitor 2 |
| chr7_+_56171081 | 0.00 |
ENSDART00000073596
ENSDART00000123273 |
ist1
|
increased sodium tolerance 1 homolog (yeast) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1903392 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 0.1 | 0.2 | GO:0003245 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.4 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.4 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.2 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 1.0 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.4 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.3 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.1 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.2 | GO:1904103 | regulation of convergent extension involved in gastrulation(GO:1904103) |
| 0.0 | 0.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.1 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.0 | 0.3 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.1 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.0 | 0.6 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 0.6 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.4 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 0.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 1.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.3 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.1 | 0.4 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.4 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0016849 | phosphorus-oxygen lyase activity(GO:0016849) |
| 0.0 | 0.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.4 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |