Project
DANIO-CODE
Navigation
Downloads
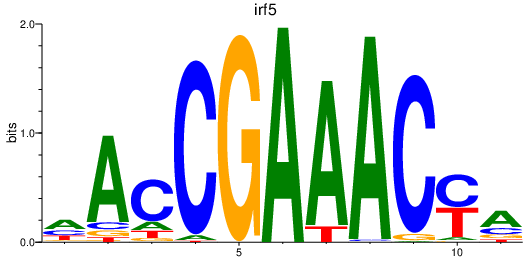
Results for irf5
Z-value: 2.42
Transcription factors associated with irf5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
irf5
|
ENSDARG00000045681 | interferon regulatory factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| irf5 | dr10_dc_chr4_-_13615927_13616074 | 0.77 | 5.5e-04 | Click! |
Activity profile of irf5 motif
Sorted Z-values of irf5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of irf5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_32937536 | 6.40 |
ENSDART00000132850
|
stard14
|
START domain containing 14 |
| chr1_-_55072271 | 5.67 |
ENSDART00000142244
|
dnajb1b
|
DnaJ (Hsp40) homolog, subfamily B, member 1b |
| chr14_-_32937496 | 5.34 |
ENSDART00000048130
|
stard14
|
START domain containing 14 |
| chr15_-_34106827 | 5.31 |
ENSDART00000163841
|
lsr
|
lipolysis stimulated lipoprotein receptor |
| chr14_+_32578103 | 5.20 |
ENSDART00000105721
|
lnx2b
|
ligand of numb-protein X 2b |
| chr7_+_68964813 | 5.20 |
ENSDART00000166258
|
marveld3
|
MARVEL domain containing 3 |
| chr22_+_24596299 | 5.17 |
ENSDART00000158303
ENSDART00000160924 |
mcoln2
|
mucolipin 2 |
| chr14_+_32578253 | 4.86 |
ENSDART00000105721
|
lnx2b
|
ligand of numb-protein X 2b |
| chr16_+_19831573 | 4.62 |
ENSDART00000135359
|
macc1
|
metastasis associated in colon cancer 1 |
| chr10_-_22180658 | 4.54 |
ENSDART00000006173
|
cldn7b
|
claudin 7b |
| chr9_-_23954640 | 4.47 |
ENSDART00000027212
|
ENSDARG00000006848
|
ENSDARG00000006848 |
| chr20_+_6545449 | 4.34 |
ENSDART00000145763
|
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr10_-_35209022 | 4.24 |
ENSDART00000063434
|
ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr10_-_22180530 | 4.08 |
ENSDART00000006173
|
cldn7b
|
claudin 7b |
| chr18_+_45669615 | 4.07 |
ENSDART00000150973
|
prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr18_+_44538877 | 4.07 |
ENSDART00000086952
|
st14a
|
suppression of tumorigenicity 14 (colon carcinoma) a |
| chr5_-_29782745 | 4.06 |
ENSDART00000125381
|
gig2o
|
grass carp reovirus (GCRV)-induced gene 2o |
| chr1_-_54505313 | 4.02 |
ENSDART00000100619
ENSDART00000163796 |
LUC7L2
|
zgc:158803 |
| chr18_+_44539099 | 3.84 |
ENSDART00000140672
|
st14a
|
suppression of tumorigenicity 14 (colon carcinoma) a |
| chr10_-_22180935 | 3.83 |
ENSDART00000006173
|
cldn7b
|
claudin 7b |
| chr5_-_28931727 | 3.76 |
ENSDART00000174697
|
arrdc1a
|
arrestin domain containing 1a |
| chr12_+_17032829 | 3.73 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr15_+_44177774 | 3.67 |
|
|
|
| chr13_-_22921202 | 3.61 |
ENSDART00000111774
|
supv3l1
|
SUV3-like helicase |
| chr10_-_33435736 | 3.58 |
ENSDART00000023509
|
ska2
|
spindle and kinetochore associated complex subunit 2 |
| chr15_+_22458649 | 3.54 |
ENSDART00000109931
|
oafa
|
OAF homolog a (Drosophila) |
| chr22_-_17663766 | 3.53 |
ENSDART00000147070
|
tjp3
|
tight junction protein 3 |
| chr13_-_37504883 | 3.53 |
ENSDART00000108855
|
si:dkey-188i13.7
|
si:dkey-188i13.7 |
| KN149966v1_+_102652 | 3.52 |
ENSDART00000160223
|
CABZ01062996.1
|
ENSDARG00000102332 |
| chr15_-_29229170 | 3.44 |
ENSDART00000138449
|
xaf1
|
XIAP associated factor 1 |
| chr20_-_22170411 | 3.41 |
ENSDART00000155568
|
BX088688.3
|
ENSDARG00000097598 |
| chr20_+_35156812 | 3.39 |
|
|
|
| chr14_-_32937341 | 3.39 |
ENSDART00000048130
|
stard14
|
START domain containing 14 |
| chr1_-_54505112 | 3.38 |
ENSDART00000100619
ENSDART00000163796 |
LUC7L2
|
zgc:158803 |
| chr15_+_44177693 | 3.31 |
|
|
|
| chr12_+_17032997 | 3.28 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr15_-_29229135 | 3.14 |
ENSDART00000138449
|
xaf1
|
XIAP associated factor 1 |
| chr6_+_30385355 | 3.14 |
ENSDART00000108943
|
shroom2a
|
shroom family member 2a |
| chr10_-_22180856 | 3.09 |
ENSDART00000006173
|
cldn7b
|
claudin 7b |
| chr15_+_44160948 | 3.04 |
ENSDART00000110060
|
zgc:165514
|
zgc:165514 |
| chr20_+_3386573 | 2.96 |
ENSDART00000175369
ENSDART00000176963 ENSDART00000176191 |
CABZ01061478.1
|
ENSDARG00000106218 |
| chr24_+_39285121 | 2.95 |
|
|
|
| chr9_-_30453581 | 2.92 |
ENSDART00000060150
|
mid1ip1a
|
MID1 interacting protein 1a |
| chr24_-_34794538 | 2.92 |
ENSDART00000171009
ENSDART00000170046 |
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr22_-_7431710 | 2.92 |
ENSDART00000170630
|
BX511034.7
|
ENSDARG00000104176 |
| chr21_+_20974216 | 2.90 |
ENSDART00000079692
|
ndr1
|
nodal-related 1 |
| chr20_+_6545194 | 2.83 |
ENSDART00000159829
|
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr9_-_23954690 | 2.74 |
ENSDART00000027212
|
ENSDARG00000006848
|
ENSDARG00000006848 |
| chr10_-_25366450 | 2.74 |
ENSDART00000123820
|
tmem135
|
transmembrane protein 135 |
| chr5_-_37655292 | 2.73 |
ENSDART00000156291
|
CT025690.1
|
ENSDARG00000096962 |
| chr6_-_1613740 | 2.70 |
ENSDART00000156305
|
trim107
|
tripartite motif containing 107 |
| chr6_-_33931989 | 2.64 |
ENSDART00000141483
|
nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr2_-_21780380 | 2.60 |
ENSDART00000144587
|
plcd1b
|
phospholipase C, delta 1b |
| chr5_+_32687543 | 2.59 |
ENSDART00000123210
|
med22
|
mediator complex subunit 22 |
| chr25_+_15983733 | 2.58 |
ENSDART00000165598
ENSDART00000061753 |
far1
|
fatty acyl CoA reductase 1 |
| chr19_-_15516318 | 2.58 |
ENSDART00000151454
|
serinc2
|
serine incorporator 2 |
| chr23_+_32015527 | 2.56 |
ENSDART00000088607
|
nemp1
|
nuclear envelope integral membrane protein 1 |
| chr6_-_10492724 | 2.53 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr10_-_22180791 | 2.44 |
ENSDART00000006173
|
cldn7b
|
claudin 7b |
| chr25_+_28419390 | 2.44 |
ENSDART00000154681
|
si:ch211-106e7.2
|
si:ch211-106e7.2 |
| chr9_-_23954761 | 2.43 |
ENSDART00000027212
|
ENSDARG00000006848
|
ENSDARG00000006848 |
| chr20_-_43846604 | 2.41 |
ENSDART00000150078
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr5_-_29782945 | 2.37 |
ENSDART00000125381
|
gig2o
|
grass carp reovirus (GCRV)-induced gene 2o |
| chr7_-_69576270 | 2.35 |
|
|
|
| chr19_+_42657913 | 2.33 |
ENSDART00000131574
ENSDART00000135436 |
jtb
|
jumping translocation breakpoint |
| chr22_+_32281360 | 2.33 |
ENSDART00000114531
ENSDART00000136125 |
rbm15b
|
RNA binding motif protein 15B |
| chr15_+_34106908 | 2.33 |
|
|
|
| chr6_+_49772891 | 2.32 |
ENSDART00000134207
|
ctsz
|
cathepsin Z |
| chr7_+_45748011 | 2.32 |
ENSDART00000170294
|
ccne1
|
cyclin E1 |
| chr15_+_25516764 | 2.30 |
ENSDART00000009545
|
pak4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr15_-_1519615 | 2.30 |
ENSDART00000129356
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr5_-_29782972 | 2.29 |
ENSDART00000125381
|
gig2o
|
grass carp reovirus (GCRV)-induced gene 2o |
| chr7_-_55879342 | 2.26 |
ENSDART00000098438
|
spg7
|
spastic paraplegia 7 |
| chr7_-_19116856 | 2.26 |
ENSDART00000165680
|
ntn4
|
netrin 4 |
| chr21_+_1719308 | 2.26 |
|
|
|
| KN149855v1_+_17122 | 2.26 |
ENSDART00000162455
ENSDART00000157851 |
CABZ01109021.1
|
ENSDARG00000100904 |
| chr10_-_20524937 | 2.24 |
|
|
|
| chr20_+_3386522 | 2.23 |
ENSDART00000175369
ENSDART00000176963 ENSDART00000176191 |
CABZ01061478.1
|
ENSDARG00000106218 |
| chr23_-_10810190 | 2.23 |
ENSDART00000140745
|
eif4e3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr7_+_45748086 | 2.22 |
ENSDART00000170294
|
ccne1
|
cyclin E1 |
| chr23_-_31339986 | 2.22 |
ENSDART00000139746
|
si:dkey-261l7.2
|
si:dkey-261l7.2 |
| chr4_+_20093671 | 2.18 |
ENSDART00000066961
|
ppp6r2a
|
protein phosphatase 6, regulatory subunit 2a |
| chr7_-_69576406 | 2.17 |
|
|
|
| chr6_-_39162916 | 2.17 |
ENSDART00000148661
|
stat2
|
signal transducer and activator of transcription 2 |
| chr24_-_34794856 | 2.15 |
ENSDART00000171009
ENSDART00000170046 |
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr5_+_44204845 | 2.15 |
ENSDART00000141198
|
ctsla
|
cathepsin La |
| chr14_+_50010976 | 2.15 |
ENSDART00000171955
|
CR855307.1
|
ENSDARG00000099628 |
| chr9_+_23954807 | 2.14 |
ENSDART00000145120
|
ENSDARG00000040445
|
ENSDARG00000040445 |
| chr25_-_27222696 | 2.12 |
ENSDART00000123590
|
si:ch211-91p5.3
|
si:ch211-91p5.3 |
| chr21_-_22507089 | 2.11 |
ENSDART00000177084
|
myo5b
|
myosin VB |
| chr5_+_44204661 | 2.10 |
ENSDART00000122288
|
ctsla
|
cathepsin La |
| chr19_+_5052459 | 2.09 |
ENSDART00000003634
|
stard3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr9_+_42656327 | 2.07 |
|
|
|
| chr9_+_38715293 | 2.06 |
ENSDART00000131846
|
osbpl11
|
oxysterol binding protein-like 11 |
| chr13_-_24778352 | 2.05 |
|
|
|
| chr18_-_44539130 | 2.04 |
|
|
|
| chr15_+_25516962 | 2.04 |
ENSDART00000165509
|
pak4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr2_+_38042821 | 2.02 |
ENSDART00000134211
ENSDART00000144868 |
hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| KN150702v1_-_111659 | 2.02 |
ENSDART00000165431
|
PPP1CC
|
protein phosphatase 1 catalytic subunit gamma |
| chr15_+_22458952 | 2.01 |
ENSDART00000109931
|
oafa
|
OAF homolog a (Drosophila) |
| chr25_+_15901398 | 2.00 |
ENSDART00000140047
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr21_-_22085596 | 1.96 |
ENSDART00000101726
|
slc35f2
|
solute carrier family 35, member F2 |
| chr7_-_69576628 | 1.92 |
|
|
|
| chr4_-_17680826 | 1.89 |
ENSDART00000066997
|
dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr7_-_29956782 | 1.89 |
ENSDART00000173514
|
znf710b
|
zinc finger protein 710b |
| chr12_+_17032963 | 1.88 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr3_-_55399158 | 1.87 |
ENSDART00000162413
|
axin2
|
axin 2 (conductin, axil) |
| chr7_-_73897752 | 1.83 |
ENSDART00000164874
|
cldnd1a
|
claudin domain containing 1a |
| chr12_-_16403282 | 1.82 |
ENSDART00000037601
|
rpp30
|
ribonuclease P/MRP 30 subunit |
| chr5_+_56772028 | 1.81 |
ENSDART00000097395
|
prpf4
|
PRP4 pre-mRNA processing factor 4 homolog (yeast) |
| chr10_-_2944190 | 1.77 |
ENSDART00000132526
|
marveld2a
|
MARVEL domain containing 2a |
| chr5_+_32687626 | 1.76 |
ENSDART00000146759
|
med22
|
mediator complex subunit 22 |
| chr21_+_37985536 | 1.74 |
ENSDART00000085728
|
klf8
|
Kruppel-like factor 8 |
| KN150384v1_+_17709 | 1.74 |
|
|
|
| chr15_+_32053944 | 1.74 |
ENSDART00000175828
|
brca2
|
breast cancer 2, early onset |
| chr14_-_25636795 | 1.73 |
|
|
|
| chr23_-_31340025 | 1.72 |
ENSDART00000139746
|
si:dkey-261l7.2
|
si:dkey-261l7.2 |
| chr6_-_1613994 | 1.72 |
ENSDART00000112118
|
trim107
|
tripartite motif containing 107 |
| chr10_-_22181059 | 1.70 |
ENSDART00000006173
|
cldn7b
|
claudin 7b |
| chr11_-_3313199 | 1.69 |
ENSDART00000002545
|
mcrs1
|
microspherule protein 1 |
| chr6_-_1613915 | 1.69 |
ENSDART00000112118
|
trim107
|
tripartite motif containing 107 |
| chr10_+_35209025 | 1.67 |
ENSDART00000126105
|
nsun5
|
NOP2/Sun domain family, member 5 |
| chr14_-_32937838 | 1.67 |
ENSDART00000132850
|
stard14
|
START domain containing 14 |
| chr10_+_110941 | 1.64 |
ENSDART00000135572
|
pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr22_+_18294579 | 1.62 |
ENSDART00000033103
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr9_+_23954669 | 1.57 |
ENSDART00000145120
|
ENSDARG00000040445
|
ENSDARG00000040445 |
| chr19_+_42658184 | 1.57 |
ENSDART00000131574
ENSDART00000135436 |
jtb
|
jumping translocation breakpoint |
| chr20_-_43846553 | 1.56 |
ENSDART00000150078
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr10_-_15921335 | 1.55 |
ENSDART00000136789
|
tjp2a
|
tight junction protein 2a (zona occludens 2) |
| chr20_-_30132167 | 1.54 |
ENSDART00000033588
|
BX248324.1
|
ENSDARG00000024870 |
| chr9_+_41657971 | 1.54 |
ENSDART00000132501
|
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr22_+_18294622 | 1.51 |
ENSDART00000033103
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr9_-_30453625 | 1.50 |
ENSDART00000060150
|
mid1ip1a
|
MID1 interacting protein 1a |
| chr5_+_44205128 | 1.50 |
ENSDART00000136965
|
ctsla
|
cathepsin La |
| chr17_+_30877631 | 1.50 |
ENSDART00000127996
|
degs2
|
delta(4)-desaturase, sphingolipid 2 |
| chr4_-_20501289 | 1.49 |
ENSDART00000132464
|
gsap
|
gamma-secretase activating protein |
| chr6_+_48349630 | 1.48 |
ENSDART00000064826
|
mov10a
|
Mov10 RISC complex RNA helicase a |
| chr5_+_57784991 | 1.47 |
ENSDART00000038602
|
slc37a2
|
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr13_+_24132617 | 1.44 |
ENSDART00000135992
|
abcb10
|
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
| chr8_+_30998992 | 1.44 |
ENSDART00000129882
|
ptgesl
|
prostaglandin E synthase 2-like |
| chr12_+_26970745 | 1.44 |
ENSDART00000153426
|
srcap
|
Snf2-related CREBBP activator protein |
| chr21_+_19283166 | 1.43 |
ENSDART00000063621
|
abraxas2a
|
abraxas 2a, BRISC complex subunit |
| chr12_-_1435708 | 1.42 |
|
|
|
| chr13_+_17564330 | 1.41 |
ENSDART00000079778
|
ifit8
|
interferon-induced protein with tetratricopeptide repeats 8 |
| chr8_+_18509772 | 1.40 |
ENSDART00000089274
|
tab3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr22_-_6970664 | 1.39 |
|
|
|
| chr10_-_2944295 | 1.38 |
ENSDART00000132526
|
marveld2a
|
MARVEL domain containing 2a |
| chr9_+_23192282 | 1.37 |
ENSDART00000021060
|
eaf2
|
ELL associated factor 2 |
| chr16_+_26727466 | 1.35 |
ENSDART00000087537
|
epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr13_-_24132195 | 1.33 |
|
|
|
| chr13_-_33348231 | 1.33 |
ENSDART00000160520
|
mad2l1bp
|
MAD2L1 binding protein |
| chr24_-_34794463 | 1.31 |
ENSDART00000171009
ENSDART00000170046 |
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr18_+_45711856 | 1.30 |
ENSDART00000077341
|
depdc7
|
DEP domain containing 7 |
| chr13_-_49886891 | 1.29 |
ENSDART00000074230
|
pkz
|
protein kinase containing Z-DNA binding domains |
| chr4_-_66538843 | 1.29 |
ENSDART00000165173
|
BX548011.1
|
ENSDARG00000103357 |
| chr19_+_1743359 | 1.28 |
ENSDART00000166744
|
dennd3a
|
DENN/MADD domain containing 3a |
| chr12_-_33704272 | 1.26 |
ENSDART00000030566
|
galk1
|
galactokinase 1 |
| chr19_-_10295398 | 1.24 |
ENSDART00000148225
|
znf865
|
zinc finger protein 865 |
| chr4_+_18525469 | 1.23 |
ENSDART00000154154
|
BX649398.1
|
ENSDARG00000097195 |
| chr7_-_20593964 | 1.21 |
ENSDART00000135509
|
fis1
|
fission, mitochondrial 1 |
| chr16_-_41716834 | 1.20 |
ENSDART00000084528
|
atp2c1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr23_-_10810264 | 1.18 |
ENSDART00000013768
|
eif4e3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr5_-_37655165 | 1.18 |
ENSDART00000156291
|
CT025690.1
|
ENSDARG00000096962 |
| chr8_+_30999182 | 1.17 |
ENSDART00000129882
|
ptgesl
|
prostaglandin E synthase 2-like |
| chr21_-_42813859 | 1.16 |
|
|
|
| chr10_+_9576840 | 1.15 |
ENSDART00000080843
|
ndufa8
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8 |
| chr12_-_33704313 | 1.15 |
ENSDART00000030566
|
galk1
|
galactokinase 1 |
| chr9_+_23954607 | 1.12 |
ENSDART00000145120
|
ENSDARG00000040445
|
ENSDARG00000040445 |
| chr14_+_623802 | 1.11 |
ENSDART00000169624
|
zgc:158257
|
zgc:158257 |
| chr5_-_12586284 | 1.10 |
ENSDART00000051664
|
ypel1
|
yippee-like 1 |
| chr25_+_8370452 | 1.09 |
ENSDART00000136530
|
fanci
|
Fanconi anemia, complementation group I |
| chr16_+_27449058 | 1.09 |
ENSDART00000132329
|
stx17
|
syntaxin 17 |
| chr3_-_23466232 | 1.04 |
ENSDART00000156897
|
ube2z
|
ubiquitin-conjugating enzyme E2Z |
| chr12_+_27151693 | 1.03 |
ENSDART00000066269
|
arl4d
|
ADP-ribosylation factor-like 4D |
| chr7_+_53952463 | 1.02 |
ENSDART00000158518
|
pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr7_-_19851362 | 1.01 |
ENSDART00000052902
|
acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr14_-_33245562 | 1.00 |
ENSDART00000023540
|
sash3
|
SAM and SH3 domain containing 3 |
| chr10_+_17414243 | 1.00 |
ENSDART00000122663
|
sppl3
|
signal peptide peptidase 3 |
| chr4_-_20501201 | 0.99 |
ENSDART00000132464
|
gsap
|
gamma-secretase activating protein |
| chr8_-_19235793 | 0.98 |
ENSDART00000036148
|
zgc:77486
|
zgc:77486 |
| chr1_-_55072375 | 0.98 |
ENSDART00000142244
|
dnajb1b
|
DnaJ (Hsp40) homolog, subfamily B, member 1b |
| chr7_+_38536733 | 0.97 |
ENSDART00000052323
|
harbi1
|
harbinger transposase derived 1 |
| chr10_+_35208897 | 0.96 |
ENSDART00000063418
|
nsun5
|
NOP2/Sun domain family, member 5 |
| chr3_-_29557642 | 0.95 |
ENSDART00000151679
|
si:ch73-233k15.2
|
si:ch73-233k15.2 |
| chr15_+_43682994 | 0.94 |
ENSDART00000168589
|
ctsc
|
cathepsin C |
| chr12_-_92728 | 0.93 |
ENSDART00000152496
|
adprm
|
ADP-ribose/CDP-alcohol diphosphatase, manganese-dependent |
| chr6_-_53333532 | 0.93 |
ENSDART00000172465
|
gnb1b
|
guanine nucleotide binding protein (G protein), beta polypeptide 1b |
| chr23_-_29627060 | 0.91 |
ENSDART00000166554
|
ube4b
|
ubiquitination factor E4B, UFD2 homolog (S. cerevisiae) |
| chr17_-_8828004 | 0.89 |
|
|
|
| chr2_-_1753013 | 0.89 |
ENSDART00000126566
|
slc22a23
|
solute carrier family 22, member 23 |
| chr4_+_20093476 | 0.88 |
ENSDART00000066961
|
ppp6r2a
|
protein phosphatase 6, regulatory subunit 2a |
| KN150702v1_-_111618 | 0.86 |
ENSDART00000165431
|
PPP1CC
|
protein phosphatase 1 catalytic subunit gamma |
| chr6_+_30385400 | 0.84 |
ENSDART00000108943
|
shroom2a
|
shroom family member 2a |
| chr17_-_24860499 | 0.84 |
ENSDART00000123147
|
zbtb8a
|
zinc finger and BTB domain containing 8A |
| chr23_-_21011877 | 0.83 |
|
|
|
| chr20_-_4115423 | 0.82 |
ENSDART00000064365
|
si:ch73-111k22.2
|
si:ch73-111k22.2 |
| chr5_+_38499102 | 0.81 |
ENSDART00000166657
|
bmp2k
|
BMP2 inducible kinase |
| chr21_+_37985567 | 0.80 |
ENSDART00000085728
|
klf8
|
Kruppel-like factor 8 |
| chr20_+_51385348 | 0.80 |
|
|
|
| chr21_-_37886693 | 0.80 |
ENSDART00000177664
|
FO704750.1
|
ENSDARG00000108937 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 19.7 | GO:0060876 | semicircular canal formation(GO:0060876) |
| 1.2 | 3.6 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.7 | 2.1 | GO:0003245 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.5 | 2.6 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.5 | 3.5 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.5 | 2.5 | GO:0034205 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.4 | 2.8 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.3 | 10.1 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.3 | 1.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.3 | 4.7 | GO:0045176 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.3 | 2.4 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.3 | 2.6 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.3 | 0.8 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.2 | 1.7 | GO:0008585 | female gonad development(GO:0008585) |
| 0.2 | 6.9 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.2 | 1.0 | GO:0030730 | regulation of sequestering of triglyceride(GO:0010889) positive regulation of sequestering of triglyceride(GO:0010890) sequestering of triglyceride(GO:0030730) |
| 0.2 | 1.2 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.2 | 1.6 | GO:1904103 | regulation of convergent extension involved in gastrulation(GO:1904103) |
| 0.2 | 0.4 | GO:0021550 | medulla oblongata development(GO:0021550) dorsal motor nucleus of vagus nerve development(GO:0021744) |
| 0.2 | 1.2 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.2 | 2.9 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.2 | 2.3 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.2 | 2.8 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.2 | 1.5 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.2 | 2.6 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.2 | 1.5 | GO:0034311 | diol metabolic process(GO:0034311) |
| 0.1 | 3.6 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 1.6 | GO:0014028 | notochord formation(GO:0014028) |
| 0.1 | 1.5 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 1.5 | GO:0060337 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 5.2 | GO:0046890 | regulation of lipid biosynthetic process(GO:0046890) |
| 0.1 | 1.0 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.1 | 0.7 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 0.3 | GO:0098586 | cytoplasmic pattern recognition receptor signaling pathway in response to virus(GO:0039528) RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) cellular response to virus(GO:0098586) |
| 0.1 | 2.7 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.1 | 2.2 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.1 | 0.6 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.1 | 1.3 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 4.5 | GO:0000082 | G1/S transition of mitotic cell cycle(GO:0000082) |
| 0.1 | 2.1 | GO:0030301 | cholesterol transport(GO:0030301) |
| 0.1 | 1.0 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 3.1 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.1 | 7.9 | GO:0008544 | epidermis development(GO:0008544) |
| 0.1 | 2.3 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 6.8 | GO:0001894 | tissue homeostasis(GO:0001894) |
| 0.1 | 3.6 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.1 | 0.8 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 2.7 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 4.3 | GO:0002573 | myeloid leukocyte differentiation(GO:0002573) |
| 0.1 | 1.4 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 3.8 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.1 | 1.4 | GO:0060029 | convergent extension involved in organogenesis(GO:0060029) |
| 0.1 | 1.8 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 0.6 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.4 | GO:0010458 | exit from mitosis(GO:0010458) |
| 0.0 | 1.9 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 1.8 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 1.4 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 1.1 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 2.6 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 4.2 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 5.3 | GO:0031016 | pancreas development(GO:0031016) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0021683 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.3 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.5 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 2.0 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 7.5 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.6 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.8 | 4.5 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.6 | 3.6 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.4 | 27.6 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.4 | 2.3 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.4 | 5.5 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.4 | 3.6 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.3 | 6.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.3 | 3.4 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.3 | 2.1 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.2 | 1.8 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 2.3 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.2 | 1.0 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.2 | 3.1 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 4.1 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 1.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 1.4 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 1.4 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 3.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.6 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 1.5 | GO:0044545 | NSL complex(GO:0044545) |
| 0.1 | 3.8 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 4.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.8 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.1 | 0.4 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 0.5 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 5.1 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.2 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 5.4 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.2 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.0 | 0.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 2.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 2.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.6 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.4 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 1.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.6 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 3.4 | GO:0019866 | mitochondrial inner membrane(GO:0005743) organelle inner membrane(GO:0019866) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.8 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.6 | 5.6 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.5 | 2.6 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.5 | 2.9 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.5 | 2.4 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.4 | 6.9 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.3 | 2.8 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.3 | 8.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.3 | 1.5 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.3 | 1.6 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.3 | 1.3 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.2 | 2.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.2 | 0.8 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 4.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.2 | 0.9 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 1.4 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 1.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 1.0 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.2 | 1.5 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.2 | 1.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 6.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.2 | 0.9 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 1.8 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 2.6 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 2.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 1.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 2.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.8 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 3.6 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 2.1 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 0.7 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.1 | 4.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 3.6 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.1 | 5.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.8 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.1 | 0.4 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 0.3 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 4.4 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 18.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.4 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 5.1 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 17.3 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.3 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 13.8 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 5.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 2.6 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 8.6 | GO:0004672 | protein kinase activity(GO:0004672) |
| 0.0 | 0.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 2.6 | GO:0008168 | methyltransferase activity(GO:0008168) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 4.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 6.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 1.5 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 2.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 2.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 2.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 2.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 1.0 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 2.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.9 | PID P73PATHWAY | p73 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.4 | 8.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.4 | 2.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.3 | 6.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 4.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 1.4 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 1.8 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 2.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 4.7 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 1.7 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 2.1 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 1.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 1.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 3.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 4.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 3.8 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |