Project
DANIO-CODE
Navigation
Downloads
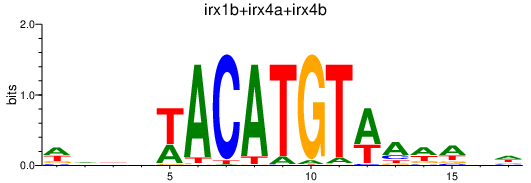
Results for irx1b+irx4a+irx4b
Z-value: 0.30
Transcription factors associated with irx1b+irx4a+irx4b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
irx4a
|
ENSDARG00000035648 | iroquois homeobox 4a |
|
irx4b
|
ENSDARG00000036051 | iroquois homeobox 4b |
|
irx1b
|
ENSDARG00000056594 | iroquois homeobox 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| irx1b | dr10_dc_chr19_-_28546417_28546461 | 0.48 | 6.3e-02 | Click! |
Activity profile of irx1b+irx4a+irx4b motif
Sorted Z-values of irx1b+irx4a+irx4b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of irx1b+irx4a+irx4b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_45155460 | 0.74 |
ENSDART00000170767
|
ved
|
ventrally expressed dharma/bozozok antagonist |
| chr13_-_43027245 | 0.52 |
ENSDART00000099601
|
ENSDARG00000068784
|
ENSDARG00000068784 |
| chr10_+_8670564 | 0.45 |
ENSDART00000129643
|
aplnrb
|
apelin receptor b |
| chr20_+_29307039 | 0.40 |
ENSDART00000152949
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr4_-_29059042 | 0.34 |
ENSDART00000169971
|
CR387997.1
|
ENSDARG00000099486 |
| chr20_+_29307142 | 0.34 |
ENSDART00000153016
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr14_-_10311677 | 0.31 |
ENSDART00000133723
|
si:dkey-92i17.2
|
si:dkey-92i17.2 |
| chr19_+_3713027 | 0.29 |
ENSDART00000125673
|
nedd9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr15_-_41777233 | 0.26 |
ENSDART00000154230
|
ftr90
|
finTRIM family, member 90 |
| chr21_-_45833636 | 0.25 |
ENSDART00000161716
|
larp1
|
La ribonucleoprotein domain family, member 1 |
| chr9_+_1964320 | 0.23 |
|
|
|
| chr15_-_14439483 | 0.23 |
ENSDART00000160675
|
dpf1
|
D4, zinc and double PHD fingers family 1 |
| chr7_-_31559907 | 0.17 |
ENSDART00000129720
|
cars
|
cysteinyl-tRNA synthetase |
| chr23_+_19775308 | 0.15 |
ENSDART00000104425
|
dnase1l1
|
deoxyribonuclease I-like 1 |
| chr23_-_45266310 | 0.14 |
|
|
|
| chr5_-_58326476 | 0.14 |
ENSDART00000033923
|
mis12
|
MIS12 kinetochore complex component |
| chr10_+_8670611 | 0.13 |
ENSDART00000129643
|
aplnrb
|
apelin receptor b |
| chr17_-_14697094 | 0.11 |
ENSDART00000154281
|
ptp4a2a
|
protein tyrosine phosphatase type IVA, member 2a |
| chr7_-_44332679 | 0.11 |
ENSDART00000073745
|
cmtm4
|
CKLF-like MARVEL transmembrane domain containing 4 |
| chr8_-_23762309 | 0.08 |
ENSDART00000141879
|
si:ch211-163l21.7
|
si:ch211-163l21.7 |
| chr1_+_966385 | 0.06 |
ENSDART00000051919
|
n6amt1
|
N-6 adenine-specific DNA methyltransferase 1 (putative) |
| chr23_-_33753484 | 0.06 |
ENSDART00000138416
|
tfcp2
|
transcription factor CP2 |
| chr7_-_18624751 | 0.05 |
ENSDART00000113593
|
mllt3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr25_+_7261050 | 0.04 |
ENSDART00000163017
|
prc1a
|
protein regulator of cytokinesis 1a |
| chr7_-_31559772 | 0.04 |
ENSDART00000052514
|
cars
|
cysteinyl-tRNA synthetase |
| chr20_-_27291090 | 0.03 |
ENSDART00000020710
ENSDART00000149024 |
btbd7
|
BTB (POZ) domain containing 7 |
| chr7_-_44332885 | 0.03 |
ENSDART00000073745
|
cmtm4
|
CKLF-like MARVEL transmembrane domain containing 4 |
| chr20_+_29307283 | 0.02 |
ENSDART00000153016
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr6_-_54049582 | 0.02 |
|
|
|
| chr10_-_28230992 | 0.01 |
ENSDART00000134491
|
med13a
|
mediator complex subunit 13a |
| chr21_-_45833527 | 0.00 |
ENSDART00000161716
|
larp1
|
La ribonucleoprotein domain family, member 1 |
| chr5_-_22090338 | 0.00 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:1904325 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) positive regulation of calcium ion transport into cytosol(GO:0010524) positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) regulation of inhibitory G-protein coupled receptor phosphorylation(GO:1904323) positive regulation of inhibitory G-protein coupled receptor phosphorylation(GO:1904325) |
| 0.1 | 0.7 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.1 | 0.3 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.0 | 0.2 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.0 | 0.2 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |