Project
DANIO-CODE
Navigation
Downloads
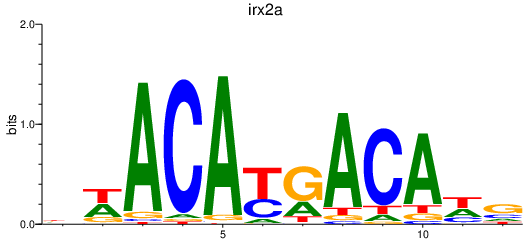
Results for irx2a
Z-value: 0.65
Transcription factors associated with irx2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
irx2a
|
ENSDARG00000001785 | iroquois homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| irx2a | dr10_dc_chr16_-_543957_544033 | -0.10 | 7.1e-01 | Click! |
Activity profile of irx2a motif
Sorted Z-values of irx2a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of irx2a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_18403762 | 0.92 |
ENSDART00000021559
|
coro1b
|
coronin, actin binding protein, 1B |
| chr21_-_32027717 | 0.64 |
ENSDART00000131651
|
ENSDARG00000073961
|
ENSDARG00000073961 |
| chr23_+_44729874 | 0.61 |
ENSDART00000056935
|
trappc1
|
trafficking protein particle complex 1 |
| chr15_+_28242729 | 0.59 |
ENSDART00000037119
|
slc46a1
|
solute carrier family 46 (folate transporter), member 1 |
| chr22_-_11049321 | 0.59 |
ENSDART00000006927
ENSDART00000170509 |
use1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr24_-_32259086 | 0.59 |
ENSDART00000048633
|
trdmt1
|
tRNA aspartic acid methyltransferase 1 |
| chr16_-_42015262 | 0.57 |
ENSDART00000136742
|
BX649388.1
|
ENSDARG00000091933 |
| chr21_-_804453 | 0.53 |
|
|
|
| chr1_+_39315964 | 0.53 |
ENSDART00000011727
|
ccdc149b
|
coiled-coil domain containing 149b |
| chr18_+_12853364 | 0.53 |
|
|
|
| chr1_-_17643893 | 0.53 |
ENSDART00000134561
|
mtnr1aa
|
melatonin receptor 1A a |
| chr8_+_25940758 | 0.52 |
ENSDART00000140626
|
ENSDARG00000061023
|
ENSDARG00000061023 |
| chr21_+_21302661 | 0.48 |
ENSDART00000137693
|
sdhaf1
|
succinate dehydrogenase complex assembly factor 1 |
| chr5_-_3323004 | 0.48 |
|
|
|
| chr5_+_31259894 | 0.47 |
ENSDART00000036235
|
iscub
|
iron-sulfur cluster assembly enzyme b |
| chr6_+_48721961 | 0.47 |
ENSDART00000164142
ENSDART00000167131 |
si:dkeyp-6a3.1
|
si:dkeyp-6a3.1 |
| chr22_+_10752209 | 0.46 |
ENSDART00000081183
|
enc3
|
ectodermal-neural cortex 3 |
| chr16_-_42225498 | 0.44 |
ENSDART00000084715
|
alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr8_+_23126151 | 0.43 |
ENSDART00000146264
|
gid8a
|
GID complex subunit 8 homolog a (S. cerevisiae) |
| chr5_-_12060314 | 0.42 |
ENSDART00000133587
|
wsb2
|
WD repeat and SOCS box containing 2 |
| chr13_-_25620394 | 0.42 |
ENSDART00000111567
|
sgpl1
|
sphingosine-1-phosphate lyase 1 |
| chr3_+_25719002 | 0.41 |
ENSDART00000007119
|
mfsd6l
|
major facilitator superfamily domain containing 6-like |
| chr11_-_16017396 | 0.41 |
ENSDART00000081062
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr16_+_32177564 | 0.41 |
|
|
|
| chr23_+_44828346 | 0.41 |
ENSDART00000140799
|
zgc:85858
|
zgc:85858 |
| chr2_+_38959335 | 0.40 |
ENSDART00000109219
|
rem2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr19_-_11397220 | 0.40 |
ENSDART00000104933
|
eepd1
|
endonuclease/exonuclease/phosphatase family domain containing 1 |
| chr7_-_51497945 | 0.40 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr21_-_32003326 | 0.39 |
ENSDART00000114964
|
zgc:165573
|
zgc:165573 |
| chr23_+_44730111 | 0.39 |
ENSDART00000177271
|
trappc1
|
trafficking protein particle complex 1 |
| chr2_-_34154845 | 0.38 |
ENSDART00000133381
|
cenpl
|
centromere protein L |
| chr16_+_40558786 | 0.38 |
ENSDART00000126129
|
nagpa
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase |
| chr1_+_11661380 | 0.38 |
ENSDART00000144475
|
clta
|
clathrin, light chain A |
| chr9_-_22014768 | 0.37 |
ENSDART00000101986
|
mrpl30
|
mitochondrial ribosomal protein L30 |
| chr24_+_31751950 | 0.36 |
ENSDART00000156493
|
CT737162.1
|
ENSDARG00000096998 |
| chr25_-_34621069 | 0.36 |
ENSDART00000123892
|
hist1h4l
|
histone 1, H4, like |
| chr22_-_28276824 | 0.35 |
ENSDART00000147686
|
si:dkey-222p3.1
|
si:dkey-222p3.1 |
| chr18_-_11426 | 0.34 |
ENSDART00000159781
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules |
| chr13_-_15662476 | 0.34 |
ENSDART00000145914
ENSDART00000010286 |
bag5
|
BCL2-associated athanogene 5 |
| chr10_-_27028132 | 0.33 |
ENSDART00000113353
|
ENSDARG00000075422
|
ENSDARG00000075422 |
| chr9_-_29687072 | 0.32 |
ENSDART00000060246
|
dnajc3a
|
DnaJ (Hsp40) homolog, subfamily C, member 3a |
| chr2_+_54911012 | 0.32 |
ENSDART00000027313
|
ndufv2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2 |
| chr18_-_14774644 | 0.32 |
ENSDART00000125979
|
tshz3a
|
teashirt zinc finger homeobox 3a |
| chr7_+_27626968 | 0.32 |
ENSDART00000178912
|
tub
|
tubby bipartite transcription factor |
| chr24_-_19573615 | 0.32 |
ENSDART00000158952
ENSDART00000109107 ENSDART00000056082 |
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr12_-_4212010 | 0.30 |
ENSDART00000059298
|
zgc:92313
|
zgc:92313 |
| chr8_+_26352980 | 0.30 |
ENSDART00000053460
ENSDART00000086763 |
nat6
hyal3
|
N-acetyltransferase 6 hyaluronoglucosaminidase 3 |
| chr7_-_24373579 | 0.30 |
ENSDART00000048921
|
rgp1
|
GP1 homolog, RAB6A GEF complex partner 1 |
| chr18_-_20619155 | 0.28 |
ENSDART00000151864
|
bcl2l13
|
BCL2-like 13 (apoptosis facilitator) |
| chr5_-_12060382 | 0.28 |
ENSDART00000133587
|
wsb2
|
WD repeat and SOCS box containing 2 |
| chr22_+_10752245 | 0.28 |
ENSDART00000050171
|
enc3
|
ectodermal-neural cortex 3 |
| chr9_-_22014861 | 0.28 |
ENSDART00000101986
|
mrpl30
|
mitochondrial ribosomal protein L30 |
| chr5_-_18393705 | 0.27 |
ENSDART00000010101
ENSDART00000099434 |
aacs
|
acetoacetyl-CoA synthetase |
| chr13_-_30011613 | 0.27 |
ENSDART00000110157
|
tysnd1
|
trypsin domain containing 1 |
| chr21_-_10962154 | 0.27 |
ENSDART00000084061
|
nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like |
| chr9_+_41223155 | 0.27 |
ENSDART00000014660
|
ormdl1
|
ORMDL sphingolipid biosynthesis regulator 1 |
| chr10_-_16069787 | 0.26 |
ENSDART00000122540
|
aldh7a1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr13_-_11838653 | 0.26 |
ENSDART00000066230
|
ARL3 (1 of many)
|
ADP ribosylation factor like GTPase 3 |
| chr25_-_34340016 | 0.26 |
ENSDART00000039485
|
gabarapl2
|
GABA(A) receptor-associated protein like 2 |
| chr17_-_30504063 | 0.25 |
ENSDART00000087111
|
itsn2b
|
intersectin 2b |
| chr7_+_13329491 | 0.25 |
ENSDART00000158797
|
ankdd1a
|
ankyrin repeat and death domain containing 1A |
| chr7_+_21006803 | 0.25 |
ENSDART00000052942
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr3_+_25718973 | 0.25 |
ENSDART00000007119
|
mfsd6l
|
major facilitator superfamily domain containing 6-like |
| chr12_+_46944395 | 0.25 |
ENSDART00000149032
|
oat
|
ornithine aminotransferase |
| chr11_+_42352106 | 0.24 |
ENSDART00000169938
|
il17rd
|
interleukin 17 receptor D |
| chr5_+_40896478 | 0.24 |
ENSDART00000039369
|
fancg
|
Fanconi anemia, complementation group G |
| chr13_-_26668719 | 0.24 |
ENSDART00000139264
|
vrk2
|
vaccinia related kinase 2 |
| chr12_-_36282538 | 0.24 |
ENSDART00000165935
|
nploc4
|
NPL4 homolog, ubiquitin recognition factor |
| chr25_-_3633631 | 0.23 |
ENSDART00000159335
ENSDART00000088077 |
zgc:158398
|
zgc:158398 |
| chr3_-_33362384 | 0.23 |
ENSDART00000020342
|
sgsm3
|
small G protein signaling modulator 3 |
| chr18_+_284210 | 0.23 |
ENSDART00000158040
|
larp6
|
La ribonucleoprotein domain family, member 6 |
| chr9_-_22014927 | 0.23 |
ENSDART00000101986
|
mrpl30
|
mitochondrial ribosomal protein L30 |
| chr13_+_11741597 | 0.23 |
|
|
|
| chr12_+_6032064 | 0.23 |
ENSDART00000142418
|
sgms1
|
sphingomyelin synthase 1 |
| chr17_+_43478181 | 0.23 |
ENSDART00000055487
|
chmp3
|
charged multivesicular body protein 3 |
| chr5_+_52199662 | 0.22 |
|
|
|
| chr16_-_40558536 | 0.22 |
ENSDART00000075718
|
ndufaf6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr12_-_46943791 | 0.22 |
ENSDART00000084557
|
lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr15_+_11798993 | 0.22 |
ENSDART00000163844
|
si:dkey-31c13.1
|
si:dkey-31c13.1 |
| chr2_+_34155114 | 0.22 |
ENSDART00000137170
|
dars2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr16_-_22218191 | 0.21 |
ENSDART00000140175
|
si:dkey-71b5.3
|
si:dkey-71b5.3 |
| chr2_-_52156537 | 0.21 |
ENSDART00000168875
|
si:ch211-9d9.1
|
si:ch211-9d9.1 |
| chr1_-_11191824 | 0.21 |
ENSDART00000163971
ENSDART00000123431 |
iqce
|
IQ motif containing E |
| chr13_-_30011574 | 0.21 |
ENSDART00000110157
|
tysnd1
|
trypsin domain containing 1 |
| chr17_+_34238753 | 0.21 |
ENSDART00000014306
|
mpp5a
|
membrane protein, palmitoylated 5a (MAGUK p55 subfamily member 5) |
| chr23_-_18454945 | 0.20 |
ENSDART00000016891
|
hsd17b10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr14_-_25688444 | 0.20 |
ENSDART00000172909
|
atox1
|
antioxidant 1 copper chaperone |
| chr5_-_12225160 | 0.20 |
ENSDART00000156408
|
lztr1
|
leucine-zipper-like transcription regulator 1 |
| chr19_-_27755498 | 0.20 |
ENSDART00000137346
|
znrd1
|
zinc ribbon domain containing 1 |
| chr3_-_23473646 | 0.19 |
ENSDART00000078423
|
atp5g1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr19_-_7522853 | 0.19 |
ENSDART00000168194
|
gabpb2a
|
GA binding protein transcription factor, beta subunit 2a |
| chr7_-_28376722 | 0.19 |
ENSDART00000150148
|
slc7a6
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 6 |
| chr25_-_34642724 | 0.19 |
ENSDART00000121963
|
hist1h4l
|
histone 1, H4, like |
| chr18_-_20618924 | 0.18 |
ENSDART00000090156
ENSDART00000151980 |
bcl2l13
|
BCL2-like 13 (apoptosis facilitator) |
| chr19_+_5562075 | 0.18 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr12_-_4212149 | 0.18 |
ENSDART00000059298
|
zgc:92313
|
zgc:92313 |
| chr7_+_23881828 | 0.18 |
ENSDART00000076735
|
lrp10
|
low density lipoprotein receptor-related protein 10 |
| chr6_-_10676890 | 0.18 |
ENSDART00000036456
|
cycsb
|
cytochrome c, somatic b |
| chr13_+_45445475 | 0.18 |
|
|
|
| chr17_-_9839620 | 0.18 |
ENSDART00000148463
|
snx6
|
sorting nexin 6 |
| chr10_-_321050 | 0.18 |
ENSDART00000165244
ENSDART00000161493 |
akt2l
|
v-akt murine thymoma viral oncogene homolog 2, like |
| chr13_-_49826526 | 0.17 |
ENSDART00000099475
|
nid1a
|
nidogen 1a |
| chr10_+_36094175 | 0.17 |
ENSDART00000164678
|
katnal1
|
katanin p60 subunit A-like 1 |
| chr8_+_47565879 | 0.17 |
|
|
|
| chr2_+_31823672 | 0.17 |
ENSDART00000086608
|
ranbp9
|
RAN binding protein 9 |
| chr16_-_22218557 | 0.17 |
ENSDART00000140175
|
si:dkey-71b5.3
|
si:dkey-71b5.3 |
| KN150226v1_+_11579 | 0.16 |
|
|
|
| chr3_-_35734943 | 0.16 |
ENSDART00000135957
|
suz12a
|
SUZ12 polycomb repressive complex 2a subunit |
| chr24_-_31967674 | 0.16 |
ENSDART00000156060
|
si:ch73-78o10.1
|
si:ch73-78o10.1 |
| chr1_-_17643855 | 0.16 |
ENSDART00000134561
|
mtnr1aa
|
melatonin receptor 1A a |
| chr14_-_49777408 | 0.16 |
ENSDART00000166463
|
cnot6b
|
CCR4-NOT transcription complex, subunit 6b |
| chr4_-_16779846 | 0.15 |
|
|
|
| chr23_-_27515964 | 0.15 |
ENSDART00000167838
|
dip2ba
|
disco-interacting protein 2 homolog Ba |
| chr18_-_13153190 | 0.15 |
ENSDART00000092648
|
tmem5
|
transmembrane protein 5 |
| chr18_-_42072998 | 0.15 |
|
|
|
| chr10_+_24530670 | 0.15 |
|
|
|
| chr7_+_24373948 | 0.15 |
ENSDART00000150233
|
gba2
|
glucosidase, beta (bile acid) 2 |
| chr5_+_40896320 | 0.15 |
ENSDART00000039369
|
fancg
|
Fanconi anemia, complementation group G |
| chr2_-_11720585 | 0.14 |
ENSDART00000019392
|
sdr16c5a
|
short chain dehydrogenase/reductase family 16C, member 5a |
| chr15_-_21003820 | 0.14 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr7_+_21651458 | 0.14 |
ENSDART00000088041
|
ufsp1
|
UFM1-specific peptidase 1 (non-functional) |
| chr3_+_57927416 | 0.14 |
ENSDART00000170144
|
pycr1a
|
pyrroline-5-carboxylate reductase 1a |
| chr13_-_15662599 | 0.14 |
ENSDART00000145914
ENSDART00000010286 |
bag5
|
BCL2-associated athanogene 5 |
| chr15_+_8791540 | 0.14 |
ENSDART00000033871
|
ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr17_+_17745119 | 0.14 |
ENSDART00000138189
ENSDART00000105013 |
alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr23_+_5632124 | 0.14 |
ENSDART00000059307
|
smpd2a
|
sphingomyelin phosphodiesterase 2a, neutral membrane (neutral sphingomyelinase) |
| chr8_-_25547089 | 0.14 |
ENSDART00000078022
|
prex1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chr20_+_31314629 | 0.14 |
ENSDART00000020252
|
pdia6
|
protein disulfide isomerase family A, member 6 |
| chr16_+_30646457 | 0.13 |
ENSDART00000058785
|
fam210ab
|
family with sequence similarity 210, member Ab |
| chr3_-_43120399 | 0.13 |
ENSDART00000166021
|
snn
|
stannin |
| chr24_+_2962301 | 0.13 |
ENSDART00000170835
|
eci2
|
enoyl-CoA delta isomerase 2 |
| chr18_-_42072902 | 0.13 |
|
|
|
| chr21_+_698978 | 0.13 |
ENSDART00000171800
ENSDART00000160956 |
oaz1b
|
ornithine decarboxylase antizyme 1b |
| chr12_+_13704914 | 0.13 |
ENSDART00000152257
|
ppp1r16a
|
protein phosphatase 1, regulatory subunit 16A |
| chr6_+_14854101 | 0.12 |
ENSDART00000087782
|
mrps9
|
mitochondrial ribosomal protein S9 |
| chr7_-_69576270 | 0.12 |
|
|
|
| chr19_+_35155362 | 0.12 |
ENSDART00000146777
ENSDART00000103276 |
fam206a
|
family with sequence similarity 206, member A |
| chr1_-_11191863 | 0.12 |
ENSDART00000163971
ENSDART00000123431 |
iqce
|
IQ motif containing E |
| chr21_+_3645203 | 0.12 |
ENSDART00000099535
|
spout1
|
SPOUT domain containing methyltransferase 1 |
| chr15_+_28242899 | 0.12 |
ENSDART00000037119
|
slc46a1
|
solute carrier family 46 (folate transporter), member 1 |
| chr20_-_33887129 | 0.12 |
ENSDART00000020183
|
fam102bb
|
family with sequence similarity 102, member B, b |
| chr2_+_47619337 | 0.11 |
ENSDART00000141974
|
acsl3b
|
acyl-CoA synthetase long-chain family member 3b |
| KN149895v1_+_73008 | 0.11 |
|
|
|
| chr4_-_28364228 | 0.11 |
ENSDART00000064219
|
trmu
|
tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase |
| chr3_+_17801981 | 0.10 |
ENSDART00000164663
|
aclya
|
ATP citrate lyase a |
| chr7_-_73632999 | 0.10 |
ENSDART00000122392
|
HIST2H2AB (1 of many)
|
histone cluster 2 H2A family member b |
| chr22_-_11618612 | 0.10 |
ENSDART00000109596
|
dpp4
|
dipeptidyl-peptidase 4 |
| chr12_-_4212095 | 0.10 |
ENSDART00000059298
|
zgc:92313
|
zgc:92313 |
| chr13_+_32505709 | 0.09 |
ENSDART00000141057
|
si:ch211-206k20.5
|
si:ch211-206k20.5 |
| chr23_+_34037058 | 0.09 |
ENSDART00000140666
|
prosc
|
pyridoxal phosphate binding protein |
| chr19_-_42933592 | 0.09 |
ENSDART00000007642
|
zgc:110239
|
zgc:110239 |
| chr9_+_8418549 | 0.09 |
ENSDART00000043067
|
zgc:171776
|
zgc:171776 |
| chr13_-_44601523 | 0.09 |
ENSDART00000074761
|
zfand3
|
zinc finger, AN1-type domain 3 |
| chr2_+_31823750 | 0.09 |
ENSDART00000086608
|
ranbp9
|
RAN binding protein 9 |
| chr21_-_37750338 | 0.09 |
ENSDART00000133585
|
fam114a2
|
family with sequence similarity 114, member A2 |
| chr14_-_28261949 | 0.09 |
ENSDART00000126095
|
insb
|
preproinsulin b |
| KN150226v1_+_11474 | 0.08 |
|
|
|
| chr6_-_10728644 | 0.08 |
ENSDART00000151102
|
notum2
|
notum pectinacetylesterase 2 |
| chr3_-_35735085 | 0.08 |
ENSDART00000135957
|
suz12a
|
SUZ12 polycomb repressive complex 2a subunit |
| chr19_+_7630847 | 0.08 |
ENSDART00000151758
|
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr24_+_14569372 | 0.07 |
ENSDART00000134475
|
gdap1
|
ganglioside induced differentiation associated protein 1 |
| chr14_+_31681774 | 0.07 |
ENSDART00000105760
|
zic6
|
zic family member 6 |
| chr1_-_44239584 | 0.07 |
ENSDART00000137216
|
tmem176
|
transmembrane protein 176 |
| chr1_+_14539897 | 0.07 |
ENSDART00000109911
|
hgsnat
|
heparan-alpha-glucosaminide N-acetyltransferase |
| chr12_-_48391342 | 0.06 |
ENSDART00000153209
|
dnajb12b
|
DnaJ (Hsp40) homolog, subfamily B, member 12b |
| chr8_-_30781902 | 0.06 |
ENSDART00000147332
|
gstt1a
|
glutathione S-transferase theta 1a |
| chr13_+_33237979 | 0.06 |
ENSDART00000158709
|
brf1a
|
BRF1, RNA polymerase III transcription initiation factor a |
| chr18_-_38289553 | 0.06 |
ENSDART00000098889
|
caprin1b
|
cell cycle associated protein 1b |
| chr22_+_1750530 | 0.06 |
|
|
|
| chr13_-_24695520 | 0.06 |
ENSDART00000142745
|
slka
|
STE20-like kinase a |
| chr5_-_43219303 | 0.06 |
ENSDART00000022481
|
mccc2
|
methylcrotonoyl-CoA carboxylase 2 (beta) |
| chr7_+_71585147 | 0.05 |
ENSDART00000161344
|
nsun6
|
NOL1/NOP2/Sun domain family, member 6 |
| chr6_+_43452686 | 0.05 |
ENSDART00000075521
|
zgc:113054
|
zgc:113054 |
| chr22_+_12570345 | 0.05 |
ENSDART00000140054
|
ENSDARG00000041602
|
ENSDARG00000041602 |
| chr11_+_38013564 | 0.05 |
ENSDART00000171496
|
CDK18
|
cyclin dependent kinase 18 |
| chr25_+_9904821 | 0.05 |
|
|
|
| chr20_-_20370397 | 0.05 |
ENSDART00000009356
|
ppp2r5ea
|
protein phosphatase 2, regulatory subunit B', epsilon isoform a |
| chr25_+_3634121 | 0.05 |
ENSDART00000153905
|
thoc5
|
THO complex 5 |
| chr2_+_31823314 | 0.05 |
ENSDART00000086608
|
ranbp9
|
RAN binding protein 9 |
| chr6_-_10728582 | 0.05 |
ENSDART00000151102
|
notum2
|
notum pectinacetylesterase 2 |
| chr25_-_22954929 | 0.04 |
ENSDART00000024633
|
dusp8a
|
dual specificity phosphatase 8a |
| chr5_+_41463969 | 0.04 |
ENSDART00000035235
|
UBB
|
si:ch211-202a12.4 |
| chr8_-_21071055 | 0.04 |
ENSDART00000131322
|
aldh9a1a.1
|
aldehyde dehydrogenase 9 family, member A1a, tandem duplicate 1 |
| chr11_+_17040226 | 0.04 |
|
|
|
| chr2_+_34154885 | 0.04 |
ENSDART00000137170
|
dars2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr13_-_32505425 | 0.04 |
ENSDART00000148040
|
matn3b
|
matrilin 3b |
| chr17_-_43437795 | 0.04 |
ENSDART00000043866
|
plk4
|
polo-like kinase 4 (Drosophila) |
| chr1_+_35919158 | 0.04 |
ENSDART00000176692
|
CR762429.1
|
ENSDARG00000108193 |
| chr15_-_24243608 | 0.03 |
ENSDART00000077980
|
pipox
|
pipecolic acid oxidase |
| chr19_-_20862383 | 0.03 |
ENSDART00000111409
ENSDART00000140711 |
tbc1d5
|
TBC1 domain family, member 5 |
| chr8_+_44933065 | 0.03 |
ENSDART00000098567
|
ENSDARG00000016545
|
ENSDARG00000016545 |
| chr5_-_66502280 | 0.03 |
ENSDART00000065264
|
cdca5
|
cell division cycle associated 5 |
| chr5_+_63716392 | 0.03 |
ENSDART00000015940
|
edf1
|
endothelial differentiation-related factor 1 |
| chr7_+_57374714 | 0.03 |
ENSDART00000110623
|
arsj
|
arylsulfatase family, member J |
| chr8_+_44933013 | 0.03 |
ENSDART00000098567
|
ENSDARG00000016545
|
ENSDARG00000016545 |
| chr8_-_21071108 | 0.03 |
ENSDART00000137838
|
aldh9a1a.1
|
aldehyde dehydrogenase 9 family, member A1a, tandem duplicate 1 |
| chr21_-_43915591 | 0.03 |
|
|
|
| chr25_-_20835675 | 0.02 |
|
|
|
| chr25_-_31952245 | 0.02 |
ENSDART00000153892
|
cep152
|
centrosomal protein 152 |
| chr23_+_24999817 | 0.02 |
ENSDART00000137486
|
klhl21
|
kelch-like family member 21 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0090083 | inclusion body assembly(GO:0070841) regulation of inclusion body assembly(GO:0090083) |
| 0.1 | 0.5 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 0.3 | GO:0090277 | positive regulation of peptide secretion(GO:0002793) positive regulation of insulin secretion(GO:0032024) positive regulation of hormone secretion(GO:0046887) positive regulation of peptide hormone secretion(GO:0090277) |
| 0.1 | 0.3 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 | 0.3 | GO:2000303 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 0.5 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 0.4 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.2 | GO:0061400 | positive regulation of transcription from RNA polymerase II promoter in response to calcium ion(GO:0061400) |
| 0.1 | 0.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 0.4 | GO:0043901 | negative regulation of multi-organism process(GO:0043901) |
| 0.0 | 0.1 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.0 | 0.1 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.0 | 0.2 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.7 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 0.1 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.4 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.5 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0051657 | maintenance of organelle location(GO:0051657) |
| 0.0 | 0.4 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.4 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.4 | GO:0030149 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.0 | 0.1 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 0.2 | GO:1901985 | positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.4 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.4 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.3 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.6 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.1 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.5 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.3 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.0 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.0 | 1.0 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.4 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.0 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0099631 | postsynaptic endocytic zone cytoplasmic component(GO:0099631) |
| 0.1 | 0.3 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.3 | GO:0034066 | RIC1-RGP1 guanyl-nucleotide exchange factor complex(GO:0034066) |
| 0.1 | 1.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:1905202 | methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.1 | 0.4 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 0.4 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.1 | 0.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.4 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 0.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.0 | 0.1 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.1 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.0 | 0.2 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.8 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 0.2 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.3 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.0 | 0.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.3 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.7 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |