Project
DANIO-CODE
Navigation
Downloads
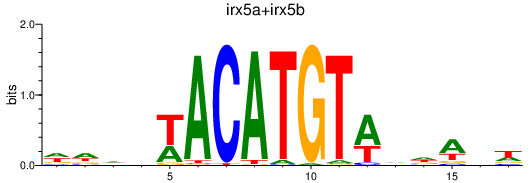
Results for irx5a+irx5b_irx3b
Z-value: 2.75
Transcription factors associated with irx5a+irx5b_irx3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
irx5a
|
ENSDARG00000034043 | iroquois homeobox 5a |
|
irx5b
|
ENSDARG00000074070 | iroquois homeobox 5b |
|
irx3b
|
ENSDARG00000031138 | iroquois homeobox 3b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| irx5a | dr10_dc_chr7_-_35438739_35438853 | -0.90 | 2.0e-06 | Click! |
| irx5b | dr10_dc_chr25_+_35602451_35602500 | -0.88 | 7.2e-06 | Click! |
| irx3b | dr10_dc_chr25_+_35901081_35901148 | -0.86 | 1.6e-05 | Click! |
Activity profile of irx5a+irx5b_irx3b motif
Sorted Z-values of irx5a+irx5b_irx3b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of irx5a+irx5b_irx3b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_33264349 | 8.64 |
|
|
|
| chr21_+_11943637 | 7.68 |
ENSDART00000141306
|
zgc:162344
|
zgc:162344 |
| chr17_+_24791024 | 7.02 |
ENSDART00000130871
|
spdya
|
speedy/RINGO cell cycle regulator family member A |
| chr23_+_46000362 | 6.61 |
ENSDART00000023944
|
lmnl3
|
lamin L3 |
| chr2_+_216779 | 6.44 |
ENSDART00000101071
|
zgc:113293
|
zgc:113293 |
| chr6_+_51713264 | 6.38 |
ENSDART00000146281
|
fam65c
|
family with sequence similarity 65, member C |
| chr17_+_24790812 | 6.25 |
ENSDART00000082251
|
spdya
|
speedy/RINGO cell cycle regulator family member A |
| chr2_+_216931 | 5.83 |
ENSDART00000101071
|
zgc:113293
|
zgc:113293 |
| chr16_-_47446494 | 5.58 |
ENSDART00000032188
|
si:dkey-256h2.1
|
si:dkey-256h2.1 |
| chr19_+_32670271 | 5.33 |
|
|
|
| chr5_+_3567992 | 5.22 |
ENSDART00000129329
|
rpain
|
RPA interacting protein |
| chr8_-_20882626 | 5.09 |
ENSDART00000025356
|
haus5
|
HAUS augmin-like complex, subunit 5 |
| chr21_+_26954898 | 4.85 |
ENSDART00000114469
|
fkbp2
|
FK506 binding protein 2 |
| chr16_-_21734502 | 4.83 |
ENSDART00000179125
|
CABZ01009710.1
|
ENSDARG00000106589 |
| chr9_+_22166405 | 4.76 |
ENSDART00000135032
|
si:dkey-57a22.11
|
si:dkey-57a22.11 |
| chr12_+_17032829 | 4.67 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr2_+_55869860 | 4.52 |
|
|
|
| chr22_+_10752209 | 4.49 |
ENSDART00000081183
|
enc3
|
ectodermal-neural cortex 3 |
| chr12_+_17032997 | 4.45 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr12_+_47448318 | 4.23 |
ENSDART00000152857
|
fmn2b
|
formin 2b |
| chr12_+_13053552 | 4.18 |
ENSDART00000124799
|
si:ch211-103b1.2
|
si:ch211-103b1.2 |
| chr7_+_38444768 | 4.05 |
ENSDART00000024590
|
syt13
|
synaptotagmin XIII |
| chr6_+_40924559 | 3.95 |
ENSDART00000133599
|
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr5_-_65564733 | 3.93 |
ENSDART00000178162
|
CABZ01049025.1
|
ENSDARG00000107190 |
| chr4_-_6407602 | 3.83 |
ENSDART00000134376
|
CR356233.2
|
ENSDARG00000093020 |
| chr3_+_17901295 | 3.79 |
ENSDART00000035531
|
mettl26
|
methyltransferase like 26 |
| chr12_+_22552867 | 3.76 |
ENSDART00000152930
|
cdca9
|
cell division cycle associated 9 |
| chr2_+_56534374 | 3.72 |
ENSDART00000113964
|
POLR2E (1 of many)
|
RNA polymerase II subunit E |
| chr3_+_33308831 | 3.69 |
ENSDART00000164322
|
gtpbp1
|
GTP binding protein 1 |
| chr6_+_33092094 | 3.65 |
ENSDART00000122242
|
pomgnt1
|
protein O-linked mannose N-acetylglucosaminyltransferase 1 (beta 1,2-) |
| chr21_-_32747592 | 3.64 |
|
|
|
| chr23_+_370986 | 3.60 |
ENSDART00000055148
|
zgc:101663
|
zgc:101663 |
| chr16_-_42236126 | 3.56 |
ENSDART00000076030
|
fbl
|
fibrillarin |
| chr6_+_40469996 | 3.53 |
ENSDART00000103868
|
zgc:152986
|
zgc:152986 |
| chr1_-_33717934 | 3.52 |
ENSDART00000083736
|
lmo7b
|
LIM domain 7b |
| chr24_+_12689711 | 3.52 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr24_+_9898743 | 3.51 |
ENSDART00000144186
|
pou6f2
|
POU class 6 homeobox 2 |
| chr14_+_7391258 | 3.44 |
ENSDART00000162363
|
brd8
|
bromodomain containing 8 |
| chr10_+_8670611 | 3.40 |
ENSDART00000129643
|
aplnrb
|
apelin receptor b |
| chr21_+_11943609 | 3.37 |
ENSDART00000155426
ENSDART00000102463 |
zgc:162344
|
zgc:162344 |
| chr19_-_7124381 | 3.37 |
ENSDART00000104845
|
tapbp.1
|
TAP binding protein (tapasin), tandem duplicate 1 |
| KN150034v1_+_1223 | 3.35 |
|
|
|
| chr10_+_37457234 | 3.32 |
ENSDART00000136554
|
akap1a
|
A kinase (PRKA) anchor protein 1a |
| chr18_+_26061521 | 3.25 |
ENSDART00000015712
|
znf710a
|
zinc finger protein 710a |
| chr10_+_24530670 | 3.24 |
|
|
|
| chr5_-_65750231 | 3.15 |
ENSDART00000160189
|
hip1rb
|
huntingtin interacting protein 1 related b |
| chr8_-_4704361 | 3.15 |
ENSDART00000064201
|
cdc45
|
CDC45 cell division cycle 45 homolog (S. cerevisiae) |
| chr20_+_31036284 | 3.07 |
ENSDART00000153344
|
sod2
|
superoxide dismutase 2, mitochondrial |
| chr23_-_31528548 | 3.06 |
|
|
|
| chr4_-_13932592 | 3.02 |
ENSDART00000067174
|
zcrb1
|
zinc finger CCHC-type and RNA binding motif 1 |
| chr4_-_7861030 | 3.01 |
ENSDART00000067339
|
mcm10
|
minichromosome maintenance 10 replication initiation factor |
| chr9_+_21548183 | 3.00 |
ENSDART00000059402
|
n6amt2
|
N-6 adenine-specific DNA methyltransferase 2 (putative) |
| chr13_+_22586873 | 2.98 |
ENSDART00000134122
|
nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr7_+_34692221 | 2.92 |
ENSDART00000085087
|
ddx28
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 28 |
| chr20_-_27291090 | 2.91 |
ENSDART00000020710
ENSDART00000149024 |
btbd7
|
BTB (POZ) domain containing 7 |
| chr24_-_24837798 | 2.91 |
ENSDART00000144961
|
slc51a
|
solute carrier family 51, alpha subunit |
| chr8_+_37716781 | 2.90 |
ENSDART00000108556
|
npm2a
|
nucleophosmin/nucleoplasmin, 2a |
| chr12_+_17032963 | 2.90 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr14_+_1226767 | 2.88 |
ENSDART00000127477
|
adad1
|
adenosine deaminase domain containing 1 (testis-specific) |
| chr22_+_1418852 | 2.87 |
|
|
|
| chr3_-_86374 | 2.86 |
ENSDART00000138302
|
zgc:110249
|
zgc:110249 |
| chr10_+_32107014 | 2.86 |
ENSDART00000137373
|
si:ch211-266i6.3
|
si:ch211-266i6.3 |
| chr2_+_15432208 | 2.85 |
ENSDART00000027171
|
f3b
|
coagulation factor IIIb |
| chr8_+_25072241 | 2.80 |
ENSDART00000143922
|
atxn7l2b
|
ataxin 7-like 2b |
| chr5_+_65491173 | 2.75 |
ENSDART00000050847
ENSDART00000172117 |
GLDC
|
glycine decarboxylase |
| chr6_+_30123156 | 2.72 |
ENSDART00000022586
|
lrrc40
|
leucine rich repeat containing 40 |
| chr5_+_33978279 | 2.72 |
ENSDART00000016314
|
gfm2
|
G elongation factor, mitochondrial 2 |
| chr19_+_7254917 | 2.72 |
ENSDART00000123934
|
hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr6_-_14841372 | 2.72 |
ENSDART00000167436
|
CU929521.1
|
ENSDARG00000097482 |
| chr5_+_65491288 | 2.71 |
ENSDART00000050847
ENSDART00000172117 |
GLDC
|
glycine decarboxylase |
| chr6_+_49902765 | 2.69 |
ENSDART00000023515
|
chmp4ba
|
charged multivesicular body protein 4Ba |
| chr16_-_21734465 | 2.69 |
|
|
|
| chr11_-_22450582 | 2.69 |
|
|
|
| chr22_+_35156074 | 2.68 |
ENSDART00000130581
|
rnf13
|
ring finger protein 13 |
| chr15_+_28242899 | 2.67 |
ENSDART00000037119
|
slc46a1
|
solute carrier family 46 (folate transporter), member 1 |
| chr3_+_28519110 | 2.66 |
ENSDART00000151116
ENSDART00000153977 |
CR792438.2
|
ENSDARG00000096409 |
| chr5_-_58326476 | 2.64 |
ENSDART00000033923
|
mis12
|
MIS12 kinetochore complex component |
| chr20_-_27291231 | 2.64 |
ENSDART00000020710
ENSDART00000149024 |
btbd7
|
BTB (POZ) domain containing 7 |
| chr10_+_37457062 | 2.63 |
ENSDART00000136554
|
akap1a
|
A kinase (PRKA) anchor protein 1a |
| chr23_-_20050419 | 2.61 |
ENSDART00000054659
|
fam58a
|
family with sequence similarity 58, member A |
| chr18_-_31126989 | 2.61 |
ENSDART00000039495
|
pdcd5
|
programmed cell death 5 |
| chr10_-_22942557 | 2.60 |
ENSDART00000131992
|
si:ch1073-143l10.2
|
si:ch1073-143l10.2 |
| chr24_+_32610430 | 2.58 |
ENSDART00000132417
|
yme1l1a
|
YME1-like 1a |
| chr22_-_510040 | 2.56 |
ENSDART00000140101
|
ccnd3
|
cyclin D3 |
| chr25_+_7261050 | 2.55 |
ENSDART00000163017
|
prc1a
|
protein regulator of cytokinesis 1a |
| chr12_-_42212994 | 2.54 |
ENSDART00000171075
|
zgc:111868
|
zgc:111868 |
| chr2_+_15432130 | 2.53 |
ENSDART00000027171
|
f3b
|
coagulation factor IIIb |
| chr8_+_3373066 | 2.53 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr11_-_6979695 | 2.52 |
ENSDART00000173242
ENSDART00000172896 ENSDART00000102493 |
zgc:173548
|
zgc:173548 |
| KN150266v1_-_68652 | 2.52 |
|
|
|
| chr20_-_22899048 | 2.51 |
ENSDART00000063609
|
fip1l1a
|
FIP1 like 1a (S. cerevisiae) |
| chr1_+_50547385 | 2.50 |
ENSDART00000132141
|
btbd3a
|
BTB (POZ) domain containing 3a |
| chr3_-_26060098 | 2.50 |
ENSDART00000136001
|
ypel3
|
yippee-like 3 |
| chr16_-_21734257 | 2.48 |
|
|
|
| chr15_+_28242729 | 2.47 |
ENSDART00000037119
|
slc46a1
|
solute carrier family 46 (folate transporter), member 1 |
| chr9_-_19358774 | 2.44 |
ENSDART00000099396
|
zgc:152951
|
zgc:152951 |
| chr8_-_53165501 | 2.44 |
ENSDART00000135982
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr20_+_25812737 | 2.44 |
ENSDART00000033436
|
cep135
|
centrosomal protein 135 |
| chr7_-_6334676 | 2.44 |
ENSDART00000081359
|
zgc:110425
|
zgc:110425 |
| chr3_-_26060047 | 2.42 |
ENSDART00000136001
|
ypel3
|
yippee-like 3 |
| chr5_-_27953681 | 2.39 |
ENSDART00000078708
|
zgc:162952
|
zgc:162952 |
| chr23_-_33753484 | 2.35 |
ENSDART00000138416
|
tfcp2
|
transcription factor CP2 |
| chr15_+_19388905 | 2.34 |
ENSDART00000149926
|
vps26b
|
VPS26 retromer complex component B |
| chr10_-_41380870 | 2.34 |
ENSDART00000160174
|
brf2
|
BRF2, RNA polymerase III transcription initiation factor |
| chr10_+_45108698 | 2.32 |
|
|
|
| chr19_-_28247275 | 2.32 |
|
|
|
| chr3_+_42395984 | 2.32 |
ENSDART00000162096
|
micall2a
|
mical-like 2a |
| chr12_-_36046366 | 2.31 |
|
|
|
| chr18_-_958923 | 2.28 |
|
|
|
| chr18_+_24490621 | 2.28 |
ENSDART00000143108
|
CR847971.1
|
ENSDARG00000091941 |
| chr18_-_44854242 | 2.27 |
ENSDART00000086823
|
srpr
|
signal recognition particle receptor (docking protein) |
| chr20_+_41351373 | 2.25 |
ENSDART00000153067
|
BX510919.1
|
ENSDARG00000096764 |
| KN150034v1_+_1158 | 2.25 |
|
|
|
| chr23_-_36207066 | 2.25 |
ENSDART00000147598
|
cbx5
|
chromobox homolog 5 (HP1 alpha homolog, Drosophila) |
| chr14_-_30604897 | 2.24 |
ENSDART00000161540
|
si:zfos-80g12.1
|
si:zfos-80g12.1 |
| chr5_-_23211957 | 2.23 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr23_-_36547458 | 2.22 |
ENSDART00000006881
|
zbtb39
|
zinc finger and BTB domain containing 39 |
| chr22_-_6532901 | 2.22 |
ENSDART00000106100
|
zgc:171490
|
zgc:171490 |
| chr17_+_22291644 | 2.22 |
ENSDART00000151929
ENSDART00000089919 ENSDART00000000804 |
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr8_+_52458939 | 2.22 |
|
|
|
| chr19_-_25565317 | 2.21 |
ENSDART00000175266
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr2_-_40857927 | 2.21 |
ENSDART00000157099
|
CT583650.1
|
ENSDARG00000096869 |
| chr7_+_26378093 | 2.19 |
ENSDART00000173823
ENSDART00000101053 |
tp53i11a
|
tumor protein p53 inducible protein 11a |
| chr9_+_21548044 | 2.19 |
ENSDART00000147619
|
n6amt2
|
N-6 adenine-specific DNA methyltransferase 2 (putative) |
| chr17_+_15780112 | 2.19 |
ENSDART00000027667
ENSDART00000161637 |
rragd
|
ras-related GTP binding D |
| chr3_+_17901095 | 2.17 |
ENSDART00000035531
|
mettl26
|
methyltransferase like 26 |
| chr22_-_5600910 | 2.17 |
ENSDART00000179579
|
CABZ01075081.1
|
ENSDARG00000106226 |
| chr1_+_9443056 | 2.16 |
ENSDART00000110698
|
rbm46
|
RNA binding motif protein 46 |
| chr6_-_54049582 | 2.15 |
|
|
|
| chr18_+_1018614 | 2.15 |
ENSDART00000161206
|
pkma
|
pyruvate kinase, muscle, a |
| chr22_+_2483845 | 2.13 |
ENSDART00000170192
|
CU570894.1
|
ENSDARG00000103886 |
| chr9_-_19358693 | 2.13 |
ENSDART00000099396
|
zgc:152951
|
zgc:152951 |
| chr3_+_17900981 | 2.12 |
ENSDART00000035531
|
mettl26
|
methyltransferase like 26 |
| chr14_-_32755454 | 2.12 |
ENSDART00000074720
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr11_-_1061904 | 2.12 |
ENSDART00000173253
ENSDART00000172765 |
CU914760.1
|
ENSDARG00000105313 |
| chr4_+_5147577 | 2.12 |
ENSDART00000067392
|
tigarb
|
tp53-induced glycolysis and apoptosis regulator b |
| chr16_+_25269188 | 2.12 |
ENSDART00000147584
|
hcst
|
hematopoietic cell signal transducer |
| chr16_+_25201900 | 2.11 |
ENSDART00000163244
|
si:ch211-261d7.6
|
si:ch211-261d7.6 |
| chr8_-_28359022 | 2.11 |
ENSDART00000062693
|
ptpn1
|
protein tyrosine phosphatase, non-receptor type 1 |
| chr5_+_68960543 | 2.10 |
ENSDART00000169013
|
arl6ip4
|
ADP-ribosylation factor-like 6 interacting protein 4 |
| chr5_+_33978304 | 2.10 |
ENSDART00000016314
|
gfm2
|
G elongation factor, mitochondrial 2 |
| chr13_-_24614722 | 2.06 |
ENSDART00000031564
|
sfr1
|
SWI5-dependent homologous recombination repair protein 1 |
| chr25_-_19388381 | 2.05 |
ENSDART00000154986
|
zgc:193812
|
zgc:193812 |
| chr20_-_32246045 | 2.04 |
ENSDART00000048537
ENSDART00000152984 |
cep57l1
|
centrosomal protein 57, like 1 |
| chr22_-_22315679 | 2.04 |
ENSDART00000033479
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr12_+_29017351 | 2.03 |
|
|
|
| chr17_+_15780156 | 2.03 |
ENSDART00000027667
ENSDART00000161637 |
rragd
|
ras-related GTP binding D |
| chr13_+_36459562 | 2.02 |
ENSDART00000030211
|
gmfb
|
glia maturation factor, beta |
| chr21_+_347169 | 2.02 |
ENSDART00000168983
|
TMEM38B
|
transmembrane protein 38B |
| chr7_+_69888002 | 2.02 |
ENSDART00000065234
|
gba3
|
glucosidase, beta, acid 3 (gene/pseudogene) |
| chr10_+_33790718 | 2.02 |
ENSDART00000161430
|
rxfp2a
|
relaxin/insulin-like family peptide receptor 2a |
| chr5_-_7591547 | 2.00 |
ENSDART00000158447
|
nipbla
|
nipped-B homolog a (Drosophila) |
| chr8_-_31044580 | 2.00 |
ENSDART00000109885
|
snrnp200
|
small nuclear ribonucleoprotein 200 (U5) |
| chr8_+_3372903 | 2.00 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr17_+_24666346 | 2.00 |
ENSDART00000146309
|
znf593
|
zinc finger protein 593 |
| chr10_-_10393735 | 1.99 |
|
|
|
| chr11_-_37613237 | 1.99 |
ENSDART00000102868
|
etnk2
|
ethanolamine kinase 2 |
| chr5_-_68765567 | 1.98 |
ENSDART00000097251
|
ENSDARG00000067564
|
ENSDARG00000067564 |
| chr23_-_31719203 | 1.97 |
ENSDART00000148122
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr17_+_14956899 | 1.97 |
|
|
|
| chr24_+_15995931 | 1.96 |
ENSDART00000105955
|
TIMM21
|
translocase of inner mitochondrial membrane 21 |
| chr15_+_17164535 | 1.96 |
ENSDART00000155350
|
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr17_+_47012113 | 1.95 |
|
|
|
| chr3_+_43073321 | 1.94 |
ENSDART00000157964
|
zc3h7a
|
zinc finger CCCH-type containing 7A |
| chr5_-_48028931 | 1.94 |
ENSDART00000031194
|
lysmd3
|
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr17_+_24919311 | 1.92 |
|
|
|
| chr24_+_12689887 | 1.91 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr20_-_25745974 | 1.90 |
ENSDART00000063137
ENSDART00000082099 |
si:dkeyp-117h8.4
|
si:dkeyp-117h8.4 |
| chr1_-_20511660 | 1.90 |
ENSDART00000087670
|
si:dkey-253i9.4
|
si:dkey-253i9.4 |
| chr17_-_29254409 | 1.89 |
ENSDART00000104219
|
rcor1
|
REST corepressor 1 |
| chr21_-_32027717 | 1.89 |
ENSDART00000131651
|
ENSDARG00000073961
|
ENSDARG00000073961 |
| chr14_-_32755189 | 1.88 |
ENSDART00000074720
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr6_-_40924459 | 1.87 |
ENSDART00000076097
|
sfi1
|
SFI1 centrin binding protein |
| chr21_-_23009910 | 1.87 |
ENSDART00000016167
|
zw10
|
zw10 kinetochore protein |
| chr6_+_21887963 | 1.86 |
ENSDART00000157796
|
cbx8b
|
chromobox homolog 8b |
| chr9_-_30174641 | 1.85 |
ENSDART00000134157
ENSDART00000089206 |
il1rapl1a
|
interleukin 1 receptor accessory protein-like 1a |
| chr2_-_47612319 | 1.84 |
ENSDART00000133615
|
BX085193.1
|
ENSDARG00000091979 |
| chr13_-_37504883 | 1.84 |
ENSDART00000108855
|
si:dkey-188i13.7
|
si:dkey-188i13.7 |
| chr20_-_38031246 | 1.84 |
ENSDART00000152989
|
angel2
|
angel homolog 2 (Drosophila) |
| chr5_+_17276357 | 1.84 |
ENSDART00000115227
|
chfr
|
checkpoint with forkhead and ring finger domains, E3 ubiquitin protein ligase |
| chr24_-_41275160 | 1.84 |
|
|
|
| chr14_-_47125993 | 1.83 |
ENSDART00000124925
|
si:ch211-235e9.8
|
si:ch211-235e9.8 |
| chr7_-_18624798 | 1.83 |
ENSDART00000113593
|
mllt3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr16_+_10373338 | 1.82 |
ENSDART00000091377
|
mrs2
|
MRS2 magnesium transporter |
| chr4_+_10605257 | 1.81 |
ENSDART00000122636
ENSDART00000037140 |
ing3
|
inhibitor of growth family, member 3 |
| chr3_+_53758069 | 1.79 |
ENSDART00000154542
|
olfm2a
|
olfactomedin 2a |
| chr8_-_4194010 | 1.79 |
|
|
|
| chr20_-_32502517 | 1.78 |
ENSDART00000153411
|
afg1lb
|
AFG1 like ATPase b |
| chr7_+_34923319 | 1.78 |
|
|
|
| chr16_-_21241981 | 1.78 |
ENSDART00000131582
|
cbx3b
|
chromobox homolog 3b |
| chr18_-_49291686 | 1.77 |
ENSDART00000174038
|
si:zfos-464b6.2
|
si:zfos-464b6.2 |
| chr20_+_14893184 | 1.76 |
ENSDART00000002463
|
tmed5
|
transmembrane p24 trafficking protein 5 |
| chr25_+_35742745 | 1.76 |
ENSDART00000152649
|
bmb
|
brambleberry |
| chr9_-_705001 | 1.76 |
ENSDART00000147092
|
cflarb
|
CASP8 and FADD-like apoptosis regulator b |
| chr14_-_8634381 | 1.76 |
ENSDART00000129030
|
zgc:153681
|
zgc:153681 |
| chr20_-_25745931 | 1.75 |
ENSDART00000063137
ENSDART00000082099 |
si:dkeyp-117h8.4
|
si:dkeyp-117h8.4 |
| chr23_-_37553481 | 1.75 |
|
|
|
| chr13_+_36459625 | 1.75 |
ENSDART00000030211
|
gmfb
|
glia maturation factor, beta |
| chr9_-_4640050 | 1.75 |
ENSDART00000171634
|
galnt13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 |
| chr10_-_22942639 | 1.74 |
ENSDART00000131992
|
si:ch1073-143l10.2
|
si:ch1073-143l10.2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.4 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 1.2 | 3.7 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 1.2 | 3.6 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 1.1 | 3.4 | GO:0071846 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) actin filament debranching(GO:0071846) |
| 1.0 | 4.8 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 1.0 | 4.8 | GO:0051177 | meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.9 | 2.7 | GO:2000191 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) regulation of organic acid transport(GO:0032890) positive regulation of organic acid transport(GO:0032892) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transport(GO:1903793) regulation of anion transmembrane transport(GO:1903959) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.8 | 3.3 | GO:0015882 | L-ascorbic acid transport(GO:0015882) L-ascorbic acid metabolic process(GO:0019852) |
| 0.8 | 3.1 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.8 | 5.5 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.7 | 4.3 | GO:0039689 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.7 | 3.4 | GO:0010524 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) positive regulation of calcium ion transport into cytosol(GO:0010524) positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) regulation of inhibitory G-protein coupled receptor phosphorylation(GO:1904323) positive regulation of inhibitory G-protein coupled receptor phosphorylation(GO:1904325) |
| 0.7 | 2.0 | GO:0030814 | regulation of cAMP metabolic process(GO:0030814) regulation of cAMP biosynthetic process(GO:0030817) regulation of adenylate cyclase activity(GO:0045761) |
| 0.6 | 3.1 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.6 | 3.7 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.6 | 1.8 | GO:0000741 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.6 | 2.9 | GO:0097009 | energy homeostasis(GO:0097009) |
| 0.6 | 3.4 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.6 | 2.2 | GO:0072334 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.5 | 6.0 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.5 | 3.2 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.5 | 1.6 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.5 | 2.1 | GO:1900543 | negative regulation of nucleotide metabolic process(GO:0045980) negative regulation of purine nucleotide metabolic process(GO:1900543) |
| 0.5 | 2.6 | GO:0034501 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.5 | 2.1 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.5 | 1.4 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.5 | 1.4 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.5 | 2.8 | GO:0010526 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.4 | 1.8 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.4 | 1.7 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.4 | 1.6 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.4 | 3.9 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.4 | 1.9 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.4 | 5.7 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.4 | 1.1 | GO:0045813 | positive regulation of Wnt signaling pathway, calcium modulating pathway(GO:0045813) |
| 0.4 | 2.9 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.3 | 1.0 | GO:0052805 | imidazole-containing compound metabolic process(GO:0052803) imidazole-containing compound catabolic process(GO:0052805) |
| 0.3 | 1.0 | GO:0042126 | nitrate metabolic process(GO:0042126) nitrate assimilation(GO:0042128) |
| 0.3 | 1.0 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.3 | 0.9 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.3 | 1.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.3 | 1.9 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.3 | 1.9 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.3 | 0.9 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.3 | 1.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.3 | 2.5 | GO:0051256 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.3 | 2.2 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.3 | 1.1 | GO:0071047 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.3 | 2.5 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.3 | 3.6 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.3 | 5.2 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.3 | 1.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.3 | 1.9 | GO:0045841 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.3 | 2.6 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.3 | 0.8 | GO:0097037 | heme export(GO:0097037) |
| 0.3 | 1.3 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.2 | 0.7 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.2 | 2.7 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.2 | 1.7 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.2 | 2.9 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 5.2 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.2 | 0.7 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.2 | 2.4 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.2 | 1.9 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.2 | 3.0 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.2 | 1.4 | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.2 | 2.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.2 | 0.9 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.2 | 1.5 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.2 | 1.1 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.2 | 3.6 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.2 | 0.6 | GO:0000388 | spliceosome conformational change to release U4 (or U4atac) and U1 (or U11)(GO:0000388) |
| 0.2 | 0.6 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.2 | 1.0 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.2 | 2.7 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.2 | 4.6 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.2 | 1.8 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.2 | 1.7 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.2 | 0.8 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.2 | 0.6 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.2 | 4.6 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.2 | 0.6 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.2 | 0.6 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.2 | 2.0 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.2 | 0.5 | GO:0051026 | chiasma assembly(GO:0051026) meiotic metaphase plate congression(GO:0051311) |
| 0.2 | 1.4 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.2 | 0.5 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.2 | 1.4 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.2 | 4.0 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.2 | 3.8 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.2 | 0.5 | GO:0000480 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.2 | 0.7 | GO:0010452 | histone H3-K36 methylation(GO:0010452) |
| 0.2 | 0.5 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.2 | 1.5 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.1 | 0.6 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 0.6 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.1 | 1.9 | GO:0030656 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of hormone metabolic process(GO:0032350) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.1 | 0.7 | GO:0045719 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 0.4 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.1 | 0.6 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.1 | 0.8 | GO:0070836 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 1.0 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.1 | 1.5 | GO:0061099 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 1.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 1.0 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.1 | 5.4 | GO:0007596 | blood coagulation(GO:0007596) hemostasis(GO:0007599) |
| 0.1 | 1.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 | 0.2 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.1 | 0.6 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) ventricular cardiac muscle cell development(GO:0055015) |
| 0.1 | 3.2 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 0.7 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 1.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 0.4 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 1.7 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.1 | 2.0 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.1 | 0.6 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.1 | 0.3 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.1 | 3.6 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.1 | 1.3 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.1 | 0.8 | GO:2000300 | regulation of synaptic vesicle transport(GO:1902803) regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.1 | 1.4 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 0.5 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.1 | 0.3 | GO:0060911 | cardiac cell fate commitment(GO:0060911) |
| 0.1 | 1.6 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 1.4 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 1.9 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.1 | 5.0 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 1.8 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.1 | 0.8 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.1 | 0.4 | GO:1903286 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 0.6 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 1.6 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.6 | GO:0060337 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 4.7 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.1 | 3.7 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 1.4 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.1 | 0.6 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.1 | 0.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 1.7 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.1 | 0.4 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.6 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 0.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 1.0 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.1 | 0.8 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.6 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.7 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 0.9 | GO:0010043 | response to zinc ion(GO:0010043) |
| 0.1 | 3.1 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 0.1 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 0.8 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 1.0 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 | 2.4 | GO:0005977 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) energy reserve metabolic process(GO:0006112) glucan metabolic process(GO:0044042) |
| 0.1 | 2.1 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 1.8 | GO:0003341 | cilium movement(GO:0003341) |
| 0.1 | 0.9 | GO:0030149 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.0 | 1.8 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 3.0 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.2 | GO:0071267 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.2 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 2.7 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.0 | 2.2 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 5.0 | GO:0051169 | nucleocytoplasmic transport(GO:0006913) nuclear transport(GO:0051169) |
| 0.0 | 1.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.3 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.0 | 3.5 | GO:0060828 | regulation of canonical Wnt signaling pathway(GO:0060828) |
| 0.0 | 0.2 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 3.7 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 2.0 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.7 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 7.8 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.7 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 1.2 | GO:0061138 | morphogenesis of a branching epithelium(GO:0061138) |
| 0.0 | 0.3 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 1.9 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.4 | GO:0046660 | female sex differentiation(GO:0046660) |
| 0.0 | 1.0 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.5 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 2.9 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.6 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 1.4 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 0.2 | GO:0010888 | negative regulation of cholesterol storage(GO:0010887) negative regulation of lipid storage(GO:0010888) |
| 0.0 | 1.3 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.0 | 0.4 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 2.1 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.5 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 1.3 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.9 | GO:0006282 | regulation of DNA repair(GO:0006282) |
| 0.0 | 1.2 | GO:0098661 | inorganic anion transmembrane transport(GO:0098661) |
| 0.0 | 1.7 | GO:0071559 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.0 | 0.1 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.0 | 0.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 2.0 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.2 | GO:0034504 | protein localization to nucleus(GO:0034504) |
| 0.0 | 0.6 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.6 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 1.5 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.0 | 0.1 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.0 | 0.3 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.0 | 0.4 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.4 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.9 | GO:0045055 | regulated exocytosis(GO:0045055) |
| 0.0 | 0.2 | GO:0048385 | regulation of retinoic acid receptor signaling pathway(GO:0048385) |
| 0.0 | 0.5 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.3 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.5 | GO:0002144 | cytosolic tRNA wobble base thiouridylase complex(GO:0002144) |
| 1.0 | 3.0 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 1.0 | 2.9 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.9 | 2.6 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.9 | 1.7 | GO:0000941 | condensed nuclear chromosome inner kinetochore(GO:0000941) |
| 0.7 | 4.8 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.6 | 4.2 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.6 | 3.9 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.5 | 1.6 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) Sec62/Sec63 complex(GO:0031207) |
| 0.4 | 3.1 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.4 | 3.6 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.4 | 1.9 | GO:0045259 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.4 | 3.7 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.3 | 1.0 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.3 | 1.0 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.3 | 1.6 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.3 | 4.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.3 | 1.3 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.3 | 0.9 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.3 | 2.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 0.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 2.7 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.2 | 2.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.2 | 6.0 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.2 | 2.5 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.2 | 0.9 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.2 | 0.5 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.2 | 1.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 3.4 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.2 | 1.0 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.2 | 2.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 5.9 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 1.9 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 1.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.0 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 3.0 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 1.0 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 4.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 2.1 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 1.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 2.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.8 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.2 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.1 | 0.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 1.8 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 5.2 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 0.4 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 0.2 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.1 | 3.9 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 0.3 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.1 | 0.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 1.1 | GO:0000177 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 0.8 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.3 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 0.6 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 1.6 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 0.3 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.1 | 1.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 2.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 1.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.6 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.4 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 2.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 7.4 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.4 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 1.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 3.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.2 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.9 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.7 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 1.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 8.5 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.4 | GO:0071010 | U2-type prespliceosome(GO:0071004) prespliceosome(GO:0071010) |
| 0.0 | 0.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.6 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 7.1 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 4.3 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 1.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.0 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 1.3 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 1.2 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 3.9 | GO:0019866 | organelle inner membrane(GO:0019866) |
| 0.0 | 0.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 4.3 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 3.4 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.1 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 0.4 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0005542 | folic acid binding(GO:0005542) |
| 1.4 | 5.6 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 1.2 | 3.6 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.9 | 2.8 | GO:0008425 | 2-polyprenyl-6-methoxy-1,4-benzoquinone methyltransferase activity(GO:0008425) quinone cofactor methyltransferase activity(GO:0030580) |
| 0.7 | 2.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.6 | 1.9 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.6 | 3.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.6 | 3.4 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.6 | 1.7 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.6 | 2.2 | GO:0005460 | UDP-glucose transmembrane transporter activity(GO:0005460) |
| 0.5 | 6.0 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.5 | 2.1 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.5 | 1.5 | GO:0035755 | cardiolipin hydrolase activity(GO:0035755) |
| 0.5 | 1.4 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.5 | 2.9 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.5 | 2.4 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.5 | 2.3 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 3 promoter sequence-specific DNA binding(GO:0001006) RNA polymerase III regulatory region DNA binding(GO:0001016) RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.4 | 4.0 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.4 | 1.7 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.4 | 3.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.3 | 1.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.3 | 2.1 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.3 | 1.0 | GO:0002094 | polyprenyltransferase activity(GO:0002094) dehydrodolichyl diphosphate synthase activity(GO:0045547) |
| 0.3 | 1.4 | GO:0072572 | poly-ADP-D-ribose binding(GO:0072572) |
| 0.3 | 1.0 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.3 | 1.0 | GO:0046857 | nitrite reductase [NAD(P)H] activity(GO:0008942) oxidoreductase activity, acting on other nitrogenous compounds as donors, with NAD or NADP as acceptor(GO:0046857) |
| 0.3 | 2.3 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.3 | 3.9 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.3 | 3.5 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.3 | 1.2 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.3 | 0.9 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.3 | 1.7 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.3 | 2.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.3 | 1.4 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.3 | 2.2 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.3 | 6.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.3 | 2.7 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.2 | 4.2 | GO:0048038 | quinone binding(GO:0048038) |
| 0.2 | 0.7 | GO:0016843 | amine-lyase activity(GO:0016843) strictosidine synthase activity(GO:0016844) |
| 0.2 | 1.4 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.2 | 1.7 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.2 | 5.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 0.7 | GO:0008251 | tRNA-specific adenosine deaminase activity(GO:0008251) |
| 0.2 | 1.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 2.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 1.6 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 1.5 | GO:0001047 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.2 | 1.9 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.2 | 3.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 1.9 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.2 | 1.0 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 2.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 0.6 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.2 | 0.8 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.2 | 0.6 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.2 | 0.9 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.2 | 0.6 | GO:0004353 | glutamate dehydrogenase (NAD+) activity(GO:0004352) glutamate dehydrogenase [NAD(P)+] activity(GO:0004353) oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.2 | 7.0 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.2 | 0.5 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.2 | 0.7 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.2 | 0.8 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.2 | 3.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 2.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 1.4 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.2 | 2.1 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.2 | 1.0 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.2 | 0.8 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 4.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 3.6 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.1 | 2.1 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.1 | 3.5 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 2.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 0.9 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.5 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 1.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.1 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.1 | 0.8 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 1.0 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 1.4 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 4.4 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 1.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 1.9 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.8 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 1.0 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 2.9 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.1 | 2.2 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 3.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.5 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.1 | 0.7 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 3.0 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 4.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.3 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.1 | 2.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 10.9 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.1 | 1.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 2.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 2.2 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.1 | 4.8 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 0.8 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 4.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 5.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 1.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.8 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.2 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 0.9 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.7 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.3 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 1.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.2 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 0.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 2.8 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.1 | 0.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 1.2 | GO:0015370 | solute:sodium symporter activity(GO:0015370) |
| 0.0 | 1.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.0 | 0.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.8 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.6 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.4 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 2.6 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.9 | GO:0022884 | protein transmembrane transporter activity(GO:0008320) macromolecule transmembrane transporter activity(GO:0022884) |
| 0.0 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 1.1 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.8 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 2.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 7.9 | GO:0016757 | transferase activity, transferring glycosyl groups(GO:0016757) |
| 0.0 | 1.2 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 1.8 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 8.5 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.3 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.6 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 9.9 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.7 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.2 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 5.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 1.5 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.4 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 1.4 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.7 | GO:0022843 | voltage-gated cation channel activity(GO:0022843) |
| 0.0 | 1.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.2 | 4.5 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.2 | 6.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 0.6 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.2 | 2.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 7.2 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 2.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 0.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 2.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 0.7 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 0.9 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 1.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 2.1 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 3.0 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 3.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 2.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.7 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 1.4 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.3 | PID ENDOTHELIN PATHWAY | Endothelins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 6.0 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.4 | 3.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.4 | 2.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.3 | 2.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.3 | 5.1 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.3 | 2.9 | REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | Genes involved in Energy dependent regulation of mTOR by LKB1-AMPK |
| 0.3 | 4.3 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.3 | 2.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 3.0 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.2 | 2.0 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 3.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 1.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 3.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 0.9 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 3.0 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.1 | 1.0 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 2.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 4.5 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.1 | 2.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 3.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 1.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 0.7 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.0 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 2.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 0.4 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 2.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.0 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 2.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.7 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.3 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |