Project
DANIO-CODE
Navigation
Downloads
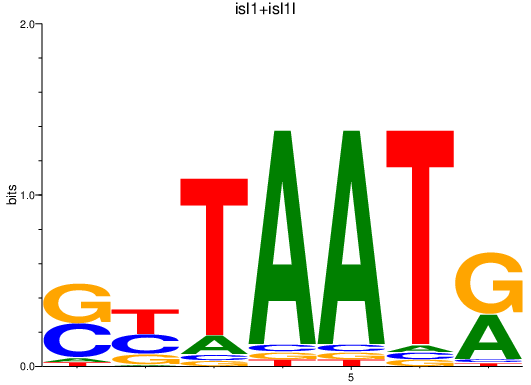
Results for isl1+isl1l
Z-value: 0.32
Transcription factors associated with isl1+isl1l
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
isl1
|
ENSDARG00000004023 | ISL LIM homeobox 1 |
|
isl1l
|
ENSDARG00000021055 | islet1, like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| isl1 | dr10_dc_chr5_-_40133824_40133903 | -0.19 | 4.7e-01 | Click! |
Activity profile of isl1+isl1l motif
Sorted Z-values of isl1+isl1l motif
Network of associatons between targets according to the STRING database.
First level regulatory network of isl1+isl1l
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_22551105 | 0.19 |
ENSDART00000123808
ENSDART00000159864 |
cdca9
|
cell division cycle associated 9 |
| chr9_-_34690993 | 0.18 |
ENSDART00000114043
|
ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr3_+_42383724 | 0.18 |
|
|
|
| chr9_-_34691159 | 0.16 |
ENSDART00000114043
|
ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr16_+_9509875 | 0.16 |
ENSDART00000162073
ENSDART00000146174 |
ice1
|
KIAA0947-like (H. sapiens) |
| chr8_-_51380691 | 0.15 |
ENSDART00000060628
|
chmp7
|
charged multivesicular body protein 7 |
| chr16_-_29452509 | 0.14 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr22_-_3279707 | 0.14 |
ENSDART00000160305
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr11_+_30400284 | 0.14 |
ENSDART00000169833
|
gb:eh507706
|
expressed sequence EH507706 |
| chr9_-_34691081 | 0.14 |
ENSDART00000114043
|
ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr12_+_26943407 | 0.13 |
ENSDART00000153054
|
fbrs
|
fibrosin |
| chr20_-_38884093 | 0.12 |
ENSDART00000153430
|
dnajc5ga
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a |
| chr3_-_58400345 | 0.12 |
ENSDART00000052179
|
cdr2a
|
cerebellar degeneration-related protein 2a |
| chr7_+_13742622 | 0.11 |
|
|
|
| chr16_+_9509747 | 0.11 |
ENSDART00000162073
ENSDART00000146174 |
ice1
|
KIAA0947-like (H. sapiens) |
| chr7_+_58910115 | 0.11 |
ENSDART00000172046
|
dok1b
|
docking protein 1b |
| chr8_+_20108592 | 0.10 |
|
|
|
| chr3_-_20912934 | 0.10 |
ENSDART00000156275
|
fam57ba
|
family with sequence similarity 57, member Ba |
| chr2_+_56016077 | 0.10 |
ENSDART00000164741
|
pgpep1
|
pyroglutamyl-peptidase I |
| chr14_-_33641145 | 0.09 |
ENSDART00000167774
|
foxa
|
forkhead box A sequence |
| chr22_-_29740737 | 0.09 |
ENSDART00000009223
|
pdcd4b
|
programmed cell death 4b |
| chr1_-_54438702 | 0.09 |
ENSDART00000142348
|
sertad2a
|
SERTA domain containing 2a |
| chr11_+_16082265 | 0.09 |
ENSDART00000081035
|
slc25a26
|
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
| chr11_+_6012910 | 0.09 |
ENSDART00000161001
|
gtpbp3
|
GTP binding protein 3 |
| chr21_+_31216418 | 0.09 |
ENSDART00000178521
|
asl
|
argininosuccinate lyase |
| chr3_-_30778382 | 0.09 |
ENSDART00000025046
|
ppp1caa
|
protein phosphatase 1, catalytic subunit, alpha isozyme a |
| chr3_-_58400454 | 0.08 |
ENSDART00000052179
|
cdr2a
|
cerebellar degeneration-related protein 2a |
| chr11_-_34909095 | 0.08 |
ENSDART00000017393
|
traip
|
TRAF-interacting protein |
| chr7_+_13742576 | 0.08 |
|
|
|
| chr2_-_56848527 | 0.08 |
ENSDART00000164330
|
CU634008.1
|
ENSDARG00000100430 |
| chr22_-_7976467 | 0.08 |
ENSDART00000162028
|
sc:d217
|
sc:d217 |
| chr2_-_32264046 | 0.08 |
ENSDART00000146452
|
fam49ba
|
family with sequence similarity 49, member Ba |
| chr9_+_19678618 | 0.08 |
ENSDART00000152034
|
si:ch211-140m22.7
|
si:ch211-140m22.7 |
| chr15_+_23721725 | 0.08 |
ENSDART00000078336
|
klc3
|
kinesin light chain 3 |
| chr14_+_19859164 | 0.08 |
ENSDART00000123434
|
fmr1
|
fragile X mental retardation 1 |
| chr1_-_7160294 | 0.08 |
ENSDART00000174972
|
BX908756.2
|
ENSDARG00000108483 |
| chr15_-_31104074 | 0.08 |
ENSDART00000157005
|
nf1a
|
neurofibromin 1a |
| chr1_-_7160099 | 0.08 |
ENSDART00000174972
|
BX908756.2
|
ENSDARG00000108483 |
| chr25_-_24150388 | 0.08 |
ENSDART00000073527
|
spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr1_+_47323244 | 0.08 |
|
|
|
| chr16_+_52779257 | 0.08 |
|
|
|
| chr8_-_985916 | 0.07 |
ENSDART00000170737
|
smyd1b
|
SET and MYND domain containing 1b |
| chr7_+_38044801 | 0.07 |
ENSDART00000160197
|
BX323855.1
|
ENSDARG00000098940 |
| chr25_-_14302207 | 0.07 |
ENSDART00000046934
|
coq9
|
coenzyme Q9 homolog (S. cerevisiae) |
| chr21_-_27376521 | 0.07 |
ENSDART00000131646
|
slc29a2
|
solute carrier family 29 (equilibrative nucleoside transporter), member 2 |
| chr17_-_29253770 | 0.07 |
ENSDART00000104219
|
rcor1
|
REST corepressor 1 |
| chr19_-_14060853 | 0.07 |
ENSDART00000161325
|
stx12
|
syntaxin 12 |
| chr6_-_19473903 | 0.07 |
ENSDART00000126186
|
psmd12
|
proteasome 26S subunit, non-ATPase 12 |
| chr15_-_20903444 | 0.07 |
ENSDART00000139551
|
aldh3a2a
|
aldehyde dehydrogenase 3 family, member A2a |
| chr5_+_56809762 | 0.07 |
ENSDART00000135520
|
si:ch211-202f5.3
|
si:ch211-202f5.3 |
| chr9_+_45804905 | 0.07 |
ENSDART00000136444
|
traf3ip1
|
TNF receptor-associated factor 3 interacting protein 1 |
| chr21_-_35806638 | 0.07 |
ENSDART00000172245
|
sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr16_-_29259838 | 0.07 |
ENSDART00000178967
|
mef2d
|
myocyte enhancer factor 2d |
| chr23_-_36349563 | 0.06 |
|
|
|
| chr9_+_37510380 | 0.06 |
ENSDART00000100622
|
slc15a2
|
solute carrier family 15 (oligopeptide transporter), member 2 |
| chr21_+_33216833 | 0.06 |
|
|
|
| chr2_-_31728072 | 0.06 |
ENSDART00000113498
|
lrrcc1
|
leucine rich repeat and coiled-coil centrosomal protein 1 |
| chr13_-_18504800 | 0.06 |
|
|
|
| chr12_-_7246994 | 0.06 |
ENSDART00000061633
|
zgc:171971
|
zgc:171971 |
| chr15_-_778461 | 0.06 |
ENSDART00000153874
|
si:dkey-7i4.19
|
si:dkey-7i4.19 |
| chr17_-_29254409 | 0.06 |
ENSDART00000104219
|
rcor1
|
REST corepressor 1 |
| chr2_-_17443642 | 0.06 |
ENSDART00000136207
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr20_-_39783396 | 0.06 |
ENSDART00000132138
ENSDART00000139252 |
BX936317.1
|
ENSDARG00000094075 |
| chr20_+_39783565 | 0.06 |
ENSDART00000153142
|
hddc2
|
HD domain containing 2 |
| chr3_-_52644257 | 0.06 |
ENSDART00000128223
|
CHCHD5
|
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr3_-_30778476 | 0.06 |
ENSDART00000025046
|
ppp1caa
|
protein phosphatase 1, catalytic subunit, alpha isozyme a |
| chr10_-_1933761 | 0.06 |
ENSDART00000101023
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr2_+_11037462 | 0.06 |
ENSDART00000081094
|
glmna
|
glomulin, FKBP associated protein a |
| chr25_-_31979408 | 0.06 |
|
|
|
| chr10_+_5191909 | 0.06 |
|
|
|
| chr2_-_11037408 | 0.06 |
ENSDART00000056034
|
rpap2
|
RNA polymerase II associated protein 2 |
| chr14_-_16816075 | 0.06 |
ENSDART00000060479
|
smtnl1
|
smoothelin-like 1 |
| chr20_+_52021000 | 0.06 |
ENSDART00000074330
|
taf1a
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A |
| chr14_-_16815986 | 0.06 |
ENSDART00000060479
|
smtnl1
|
smoothelin-like 1 |
| chr25_+_31979838 | 0.06 |
ENSDART00000012600
|
galk2
|
galactokinase 2 |
| chr3_-_39208943 | 0.06 |
|
|
|
| chr8_-_6899611 | 0.06 |
ENSDART00000139545
|
wdr13
|
WD repeat domain 13 |
| chr16_+_45370954 | 0.06 |
ENSDART00000147343
|
dnase2
|
deoxyribonuclease II, lysosomal |
| chr3_-_8608620 | 0.06 |
|
|
|
| chr18_-_50439653 | 0.06 |
ENSDART00000151038
|
CU862078.1
|
ENSDARG00000096262 |
| chr14_+_23420053 | 0.06 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr17_+_44327269 | 0.06 |
ENSDART00000045882
|
ap5m1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr23_-_9930087 | 0.06 |
|
|
|
| chr1_-_43026362 | 0.06 |
ENSDART00000132542
|
bdh2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr21_+_26486084 | 0.06 |
ENSDART00000134617
|
adssl
|
adenylosuccinate synthase, like |
| chr21_-_14276856 | 0.06 |
|
|
|
| chr21_-_4530904 | 0.06 |
ENSDART00000031425
|
zgc:55582
|
zgc:55582 |
| chr14_-_26113593 | 0.05 |
ENSDART00000020582
|
tmed9
|
transmembrane p24 trafficking protein 9 |
| chr1_+_47323043 | 0.05 |
|
|
|
| chr6_-_3347207 | 0.05 |
ENSDART00000151416
|
si:ch1073-342h5.2
|
si:ch1073-342h5.2 |
| chr9_-_43015354 | 0.05 |
|
|
|
| chr18_+_5005303 | 0.05 |
|
|
|
| chr23_-_36469779 | 0.05 |
ENSDART00000168246
|
spryd3
|
SPRY domain containing 3 |
| chr13_-_42410700 | 0.05 |
ENSDART00000134533
|
btaf1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated |
| chr8_-_51380846 | 0.05 |
ENSDART00000060628
|
chmp7
|
charged multivesicular body protein 7 |
| chr11_+_44696399 | 0.05 |
ENSDART00000158188
|
mgea5l
|
meningioma expressed antigen 5 (hyaluronidase) like |
| chr19_-_33326993 | 0.05 |
ENSDART00000124246
|
mtdha
|
metadherin a |
| chr23_-_37010664 | 0.05 |
ENSDART00000102886
|
zgc:193690
|
zgc:193690 |
| chr7_+_24543671 | 0.05 |
ENSDART00000173581
|
si:ch211-226l4.6
|
si:ch211-226l4.6 |
| chr18_+_2250457 | 0.05 |
|
|
|
| chr24_-_32750010 | 0.05 |
ENSDART00000038364
|
ca2
|
carbonic anhydrase II |
| chr6_-_19432410 | 0.05 |
ENSDART00000006843
|
cacng1a
|
calcium channel, voltage-dependent, gamma subunit 1a |
| chr22_-_29740287 | 0.05 |
ENSDART00000166002
|
pdcd4b
|
programmed cell death 4b |
| chr7_+_55797154 | 0.05 |
ENSDART00000098453
|
cdh15
|
cadherin 15, type 1, M-cadherin (myotubule) |
| chr18_-_35861412 | 0.05 |
ENSDART00000088488
|
opa3
|
optic atrophy 3 |
| chr17_-_33764220 | 0.05 |
ENSDART00000043651
|
dnal1
|
dynein, axonemal, light chain 1 |
| chr25_-_21718647 | 0.05 |
ENSDART00000152014
ENSDART00000177876 |
ENSDARG00000051873
|
ENSDARG00000051873 |
| chr2_-_2509717 | 0.05 |
ENSDART00000082143
ENSDART00000158864 |
prkacbb
|
protein kinase, cAMP-dependent, catalytic, beta b |
| chr3_-_39912816 | 0.05 |
ENSDART00000102540
|
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr3_+_53518563 | 0.05 |
ENSDART00000170461
|
col5a3a
|
collagen, type V, alpha 3a |
| chr25_+_35408674 | 0.05 |
ENSDART00000022437
|
gpia
|
glucose-6-phosphate isomerase a |
| chr18_-_40518772 | 0.05 |
ENSDART00000021372
|
chrna5
|
cholinergic receptor, nicotinic, alpha 5 |
| chr18_-_5004815 | 0.04 |
|
|
|
| chr21_+_26485851 | 0.04 |
ENSDART00000134617
|
adssl
|
adenylosuccinate synthase, like |
| chr23_+_3788435 | 0.04 |
ENSDART00000141782
|
smim29
|
small integral membrane protein 29 |
| chr9_-_19154264 | 0.04 |
ENSDART00000081878
|
pou1f1
|
POU class 1 homeobox 1 |
| chr22_-_16154562 | 0.04 |
ENSDART00000090390
|
vcam1b
|
vascular cell adhesion molecule 1b |
| chr9_-_25437880 | 0.04 |
|
|
|
| chr20_+_25812737 | 0.04 |
ENSDART00000033436
|
cep135
|
centrosomal protein 135 |
| chr24_-_23639325 | 0.04 |
ENSDART00000090368
|
sgk3
|
serum/glucocorticoid regulated kinase family, member 3 |
| KN149859v1_+_8756 | 0.04 |
ENSDART00000157687
|
ENSDARG00000103399
|
ENSDARG00000103399 |
| chr21_+_2480139 | 0.04 |
ENSDART00000162351
|
hmgcrb
|
3-hydroxy-3-methylglutaryl-CoA reductase b |
| chr10_+_15106739 | 0.04 |
ENSDART00000128967
|
parm1
|
prostate androgen-regulated mucin-like protein 1 |
| chr18_-_47026460 | 0.04 |
ENSDART00000108574
|
gramd1bb
|
GRAM domain containing 1Bb |
| chr23_+_20505072 | 0.04 |
ENSDART00000132920
|
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr15_+_27454643 | 0.04 |
ENSDART00000164887
ENSDART00000018603 |
tbx4
|
T-box 4 |
| chr22_-_14090538 | 0.04 |
ENSDART00000105717
|
aox5
|
aldehyde oxidase 5 |
| chr2_+_1596524 | 0.04 |
ENSDART00000163310
|
exosc4
|
exosome component 4 |
| chr5_+_21673232 | 0.04 |
ENSDART00000131223
|
fdx1b
|
ferredoxin 1b |
| chr2_-_32755182 | 0.04 |
ENSDART00000041146
|
nrbp2a
|
nuclear receptor binding protein 2a |
| chr10_+_2656840 | 0.04 |
|
|
|
| chr14_-_30536363 | 0.04 |
ENSDART00000147597
|
ubl3b
|
ubiquitin-like 3b |
| chr21_-_42872362 | 0.04 |
ENSDART00000171089
ENSDART00000160998 |
stk10
|
serine/threonine kinase 10 |
| chr7_+_26977165 | 0.04 |
|
|
|
| chr3_+_3826209 | 0.04 |
ENSDART00000150931
|
cdc42ep1b
|
CDC42 effector protein (Rho GTPase binding) 1b |
| chr16_+_50355723 | 0.04 |
ENSDART00000166664
|
ENSDARG00000102916
|
ENSDARG00000102916 |
| chr11_+_44696462 | 0.04 |
ENSDART00000158188
|
mgea5l
|
meningioma expressed antigen 5 (hyaluronidase) like |
| chr8_-_45997341 | 0.04 |
ENSDART00000168567
|
fam160b2
|
family with sequence similarity 160, member B2 |
| chr2_+_52472815 | 0.04 |
ENSDART00000146418
|
shda
|
Src homology 2 domain containing transforming protein D, a |
| chr22_-_29740663 | 0.04 |
ENSDART00000009223
|
pdcd4b
|
programmed cell death 4b |
| chr10_-_22875997 | 0.03 |
ENSDART00000079454
|
vamp2
|
vesicle-associated membrane protein 2 |
| chr17_-_29253918 | 0.03 |
ENSDART00000104219
|
rcor1
|
REST corepressor 1 |
| chr2_-_57785550 | 0.03 |
ENSDART00000140060
|
si:dkeyp-68b7.7
|
si:dkeyp-68b7.7 |
| chr13_-_42274486 | 0.03 |
ENSDART00000043069
|
march5
|
membrane-associated ring finger (C3HC4) 5 |
| chr9_+_41657971 | 0.03 |
ENSDART00000132501
|
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr18_-_17158316 | 0.03 |
ENSDART00000141873
|
zc3h18
|
zinc finger CCCH-type containing 18 |
| chr25_+_18910528 | 0.03 |
ENSDART00000104420
|
samm50
|
SAMM50 sorting and assembly machinery component |
| chr4_-_27907868 | 0.03 |
ENSDART00000066924
|
tbc1d22a
|
TBC1 domain family, member 22a |
| chr6_+_8362892 | 0.03 |
ENSDART00000032118
|
ube2g2
|
ubiquitin-conjugating enzyme E2G 2 (UBC7 homolog, yeast) |
| chr16_+_5377008 | 0.03 |
ENSDART00000133972
|
plecb
|
plectin b |
| KN150604v1_-_7233 | 0.03 |
|
|
|
| chr15_+_35076414 | 0.03 |
ENSDART00000165210
|
zgc:66024
|
zgc:66024 |
| chr7_-_49322116 | 0.03 |
ENSDART00000174161
|
brsk2b
|
BR serine/threonine kinase 2b |
| chr8_+_3373066 | 0.03 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr12_+_13167838 | 0.03 |
ENSDART00000160664
|
ppp1cab
|
protein phosphatase 1, catalytic subunit, alpha isozyme b |
| chr9_-_35062828 | 0.03 |
ENSDART00000026378
|
slc25a6
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 |
| chr19_+_37593325 | 0.03 |
|
|
|
| chr22_+_35229139 | 0.03 |
ENSDART00000061315
ENSDART00000146430 |
tsc22d2
|
TSC22 domain family 2 |
| chr2_-_44329092 | 0.03 |
|
|
|
| chr22_-_3661745 | 0.03 |
ENSDART00000153634
|
FP565432.1
|
ENSDARG00000097145 |
| chr7_-_9628384 | 0.03 |
ENSDART00000006343
|
asb7
|
ankyrin repeat and SOCS box containing 7 |
| chr3_-_29837017 | 0.03 |
ENSDART00000139310
|
bcat2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr20_-_45806178 | 0.03 |
ENSDART00000027482
|
gpcpd1
|
glycerophosphocholine phosphodiesterase 1 |
| chr13_-_18222908 | 0.03 |
ENSDART00000178407
|
si:dkey-228d14.5
|
si:dkey-228d14.5 |
| chr20_-_45868388 | 0.03 |
ENSDART00000062092
|
trmt6
|
tRNA methyltransferase 6 homolog (S. cerevisiae) |
| chr2_+_58661357 | 0.03 |
|
|
|
| chr5_-_32680882 | 0.03 |
ENSDART00000138116
|
surf6
|
surfeit 6 |
| chr5_+_50223133 | 0.03 |
ENSDART00000128402
|
nol6
|
nucleolar protein 6 (RNA-associated) |
| chr1_-_10391257 | 0.03 |
ENSDART00000102903
|
dmd
|
dystrophin |
| chr7_+_67206056 | 0.03 |
ENSDART00000162553
|
kars
|
lysyl-tRNA synthetase |
| chr5_+_69105916 | 0.03 |
ENSDART00000073671
|
ythdc1
|
YTH domain containing 1 |
| chr12_-_13167362 | 0.03 |
ENSDART00000152670
|
pelo
|
pelota mRNA surveillance and ribosome rescue factor |
| chr2_+_21698627 | 0.03 |
ENSDART00000133228
|
ctdsplb
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like b |
| chr4_-_19027117 | 0.03 |
ENSDART00000166160
|
si:dkey-31f5.11
|
si:dkey-31f5.11 |
| chr3_-_33789615 | 0.03 |
|
|
|
| chr17_-_29254052 | 0.03 |
ENSDART00000104219
|
rcor1
|
REST corepressor 1 |
| chr16_-_22218557 | 0.03 |
ENSDART00000140175
|
si:dkey-71b5.3
|
si:dkey-71b5.3 |
| chr16_+_17706214 | 0.03 |
|
|
|
| chr22_+_31073990 | 0.03 |
ENSDART00000111561
|
zmp:0000000735
|
zmp:0000000735 |
| chr14_+_21831841 | 0.03 |
ENSDART00000132514
|
ccng1
|
cyclin G1 |
| chr1_-_44621977 | 0.03 |
|
|
|
| chr11_+_25165838 | 0.03 |
ENSDART00000089120
|
adnpa
|
activity-dependent neuroprotector homeobox a |
| chr21_-_21142020 | 0.02 |
ENSDART00000079678
|
ftsj1
|
FtsJ RNA methyltransferase homolog 1 (E. coli) |
| chr23_-_25853331 | 0.02 |
ENSDART00000110670
|
ENSDARG00000078574
|
ENSDARG00000078574 |
| chr25_+_3232912 | 0.02 |
ENSDART00000104877
ENSDART00000170765 ENSDART00000158571 |
chchd3b
|
coiled-coil-helix-coiled-coil-helix domain containing 3b |
| chr6_-_22862617 | 0.02 |
ENSDART00000160887
|
gga3
|
ggolgi-associated, gamma adaptin ear containing, ARF binding protein 3 |
| chr25_+_29687993 | 0.02 |
ENSDART00000152705
|
api5
|
apoptosis inhibitor 5 |
| chr12_+_31523411 | 0.02 |
ENSDART00000153129
|
dnmbp
|
dynamin binding protein |
| chr12_-_45496246 | 0.02 |
ENSDART00000176229
ENSDART00000175457 |
FO818685.1
|
ENSDARG00000107895 |
| chr3_+_34856462 | 0.02 |
ENSDART00000121981
|
smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr19_+_42863138 | 0.02 |
ENSDART00000076938
|
pogza
|
pogo transposable element derived with ZNF domain a |
| chr21_+_26485814 | 0.02 |
ENSDART00000134617
|
adssl
|
adenylosuccinate synthase, like |
| chr23_+_20505319 | 0.02 |
ENSDART00000140219
|
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr9_+_38814858 | 0.02 |
ENSDART00000114728
|
znf148
|
zinc finger protein 148 |
| chr13_-_30011574 | 0.02 |
ENSDART00000110157
|
tysnd1
|
trypsin domain containing 1 |
| chr7_+_55797053 | 0.02 |
ENSDART00000127046
|
cdh15
|
cadherin 15, type 1, M-cadherin (myotubule) |
| chr1_-_7159912 | 0.02 |
ENSDART00000174972
|
BX908756.2
|
ENSDARG00000108483 |
| chr19_-_6274237 | 0.02 |
ENSDART00000151405
|
erf
|
Ets2 repressor factor |
| chr10_-_24796373 | 0.02 |
ENSDART00000064463
|
timm10b
|
translocase of inner mitochondrial membrane 10 homolog B (yeast) |
| chr23_-_37010705 | 0.02 |
ENSDART00000134461
|
zgc:193690
|
zgc:193690 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070424 | peptidoglycan transport(GO:0015835) nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway(GO:0035872) nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.0 | 0.1 | GO:0072045 | convergent extension involved in nephron morphogenesis(GO:0072045) |
| 0.0 | 0.1 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.0 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.1 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.0 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.2 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.1 | GO:0006921 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.2 | GO:1900052 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.0 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.0 | 0.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.1 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.0 | 0.1 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.0 | 0.1 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.0 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.0 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |