Project
DANIO-CODE
Navigation
Downloads
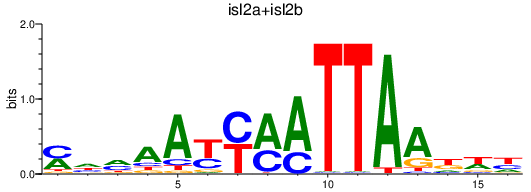
Results for isl2a+isl2b
Z-value: 0.68
Transcription factors associated with isl2a+isl2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
isl2a
|
ENSDARG00000003971 | ISL LIM homeobox 2a |
|
isl2b
|
ENSDARG00000053499 | ISL LIM homeobox 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| isl2a | dr10_dc_chr25_-_32344881_32344940 | 0.08 | 7.7e-01 | Click! |
Activity profile of isl2a+isl2b motif
Sorted Z-values of isl2a+isl2b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of isl2a+isl2b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_2677200 | 1.45 |
ENSDART00000063320
|
cldne
|
claudin e |
| chr6_-_37444877 | 1.29 |
ENSDART00000138351
|
cth
|
cystathionase (cystathionine gamma-lyase) |
| chr7_-_17129072 | 1.00 |
|
|
|
| chr7_+_38895198 | 0.94 |
ENSDART00000052318
|
mdka
|
midkine a |
| chr25_-_22821132 | 0.94 |
|
|
|
| chr20_-_475417 | 0.93 |
ENSDART00000032212
|
fynrk
|
fyn-related Src family tyrosine kinase |
| chr1_+_13779755 | 0.90 |
|
|
|
| chr7_-_48392300 | 0.89 |
|
|
|
| chr22_-_36561247 | 0.75 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr17_+_15425559 | 0.72 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr16_+_54350976 | 0.72 |
ENSDART00000172622
ENSDART00000020033 |
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr16_+_4825019 | 0.72 |
ENSDART00000167665
|
LIN28A
|
lin-28 homolog A |
| chr18_+_17548213 | 0.71 |
ENSDART00000144960
|
nup93
|
nucleoporin 93 |
| chr10_+_29964045 | 0.70 |
ENSDART00000118838
|
SNORD14
|
Small nucleolar RNA SNORD14 |
| chr5_-_19357961 | 0.67 |
ENSDART00000088904
ENSDART00000163701 |
gltpa
|
glycolipid transfer protein a |
| chr9_+_2523927 | 0.65 |
ENSDART00000166326
|
si:ch73-167c12.2
|
si:ch73-167c12.2 |
| chr17_+_1799209 | 0.64 |
ENSDART00000112183
|
cep170b
|
centrosomal protein 170B |
| chr3_-_26657213 | 0.59 |
|
|
|
| chr19_-_5851328 | 0.57 |
ENSDART00000133106
|
si:ch211-264f5.6
|
si:ch211-264f5.6 |
| chr11_-_44724371 | 0.55 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr24_+_35500964 | 0.55 |
ENSDART00000058571
|
snai2
|
snail family zinc finger 2 |
| chr12_-_22438379 | 0.55 |
ENSDART00000177715
|
CR925753.1
|
ENSDARG00000108493 |
| chr13_-_36265504 | 0.54 |
ENSDART00000140243
|
actn1
|
actinin, alpha 1 |
| chr4_-_73485204 | 0.53 |
ENSDART00000122898
|
phf21b
|
PHD finger protein 21B |
| chr17_-_16414629 | 0.51 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr14_-_24094142 | 0.49 |
ENSDART00000126894
|
fam13b
|
family with sequence similarity 13, member B |
| chr21_-_8103907 | 0.48 |
ENSDART00000171027
|
OLFML2A
|
olfactomedin like 2A |
| chr16_-_48458505 | 0.48 |
ENSDART00000159372
|
eif3ha
|
eukaryotic translation initiation factor 3, subunit H, a |
| chr10_+_29963998 | 0.46 |
ENSDART00000118838
|
SNORD14
|
Small nucleolar RNA SNORD14 |
| chr12_-_4497094 | 0.45 |
ENSDART00000163651
|
trpm4b.2
|
transient receptor potential cation channel, subfamily M, member 4b, transient receptor potential cation channel, subfamily M, member 4b, tandem duplicate 2 |
| chr2_-_57133471 | 0.44 |
|
|
|
| chr10_-_43924675 | 0.44 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr3_-_19218660 | 0.44 |
ENSDART00000132987
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr14_-_6831373 | 0.43 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr20_+_41123859 | 0.42 |
ENSDART00000146052
|
man1a1
|
mannosidase, alpha, class 1A, member 1 |
| chr19_+_38834972 | 0.42 |
ENSDART00000140198
ENSDART00000033037 ENSDART00000035093 |
col9a2
|
procollagen, type IX, alpha 2 |
| chr6_-_53227526 | 0.42 |
ENSDART00000160955
|
slc38a3b
|
solute carrier family 38, member 3b |
| chr24_-_14447773 | 0.42 |
|
|
|
| chr8_+_28362311 | 0.41 |
ENSDART00000062682
|
adipor1b
|
adiponectin receptor 1b |
| chr14_-_33604914 | 0.41 |
ENSDART00000168546
|
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr23_-_43595189 | 0.41 |
|
|
|
| chr13_+_516287 | 0.41 |
ENSDART00000172469
ENSDART00000168523 ENSDART00000006892 |
tp53bp2a
|
tumor protein p53 binding protein, 2a |
| chr13_+_516546 | 0.37 |
ENSDART00000172469
ENSDART00000168523 ENSDART00000006892 |
tp53bp2a
|
tumor protein p53 binding protein, 2a |
| chr16_-_17164459 | 0.37 |
ENSDART00000178443
|
tnfrsf1a
|
tumor necrosis factor receptor superfamily, member 1a |
| chr16_-_7876036 | 0.34 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr22_-_23518719 | 0.34 |
ENSDART00000161253
|
crb1
|
crumbs family member 1, photoreceptor morphogenesis associated |
| chr17_-_11312185 | 0.33 |
ENSDART00000091159
|
adpgk2
|
ADP-dependent glucokinase 2 |
| chr17_+_43605118 | 0.33 |
ENSDART00000156271
|
cfap99
|
cilia and flagella associated protein 99 |
| chr10_-_17264097 | 0.32 |
ENSDART00000134059
|
depdc5
|
DEP domain containing 5 |
| chr20_-_54206786 | 0.32 |
ENSDART00000004756
|
hsp90aa1.1
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 1 |
| chr19_+_16111610 | 0.32 |
ENSDART00000131319
|
ctps1a
|
CTP synthase 1a |
| chr4_-_17680826 | 0.31 |
ENSDART00000066997
|
dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr20_+_22900165 | 0.31 |
ENSDART00000058527
|
scfd2
|
sec1 family domain containing 2 |
| chr13_-_36265476 | 0.31 |
ENSDART00000133740
ENSDART00000100217 |
actn1
|
actinin, alpha 1 |
| chr7_+_58007815 | 0.31 |
ENSDART00000179306
|
CR354540.2
|
ENSDARG00000106881 |
| chr20_-_475321 | 0.30 |
ENSDART00000032212
|
fynrk
|
fyn-related Src family tyrosine kinase |
| chr10_+_29964164 | 0.29 |
ENSDART00000118838
|
SNORD14
|
Small nucleolar RNA SNORD14 |
| chr12_+_19159234 | 0.28 |
ENSDART00000153428
|
tnrc6b
|
trinucleotide repeat containing 6b |
| chr9_-_32942783 | 0.26 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr12_-_28248133 | 0.26 |
ENSDART00000016283
|
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr21_+_25199691 | 0.24 |
ENSDART00000168140
ENSDART00000112783 |
tmem45b
|
transmembrane protein 45B |
| chr9_-_34691159 | 0.24 |
ENSDART00000114043
|
ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr5_-_9162684 | 0.23 |
|
|
|
| chr21_+_13196717 | 0.23 |
ENSDART00000163767
|
adora2ab
|
adenosine A2a receptor b |
| chr14_-_6831308 | 0.23 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr25_-_20933295 | 0.23 |
ENSDART00000138985
ENSDART00000046298 |
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr13_+_516496 | 0.23 |
ENSDART00000172469
ENSDART00000168523 ENSDART00000006892 |
tp53bp2a
|
tumor protein p53 binding protein, 2a |
| chr8_-_52728470 | 0.22 |
|
|
|
| chr7_-_5253397 | 0.22 |
ENSDART00000033316
|
vangl2
|
VANGL planar cell polarity protein 2 |
| chr18_+_25743054 | 0.22 |
|
|
|
| chr22_-_4867538 | 0.22 |
ENSDART00000081801
|
ncl1
|
nicalin |
| chr1_+_52157637 | 0.22 |
ENSDART00000123972
|
smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr23_+_33792005 | 0.21 |
ENSDART00000024695
|
dazap2
|
DAZ associated protein 2 |
| chr2_-_48203102 | 0.21 |
|
|
|
| chr16_+_5492632 | 0.20 |
|
|
|
| chr18_+_17594381 | 0.19 |
ENSDART00000010998
|
slc12a3
|
solute carrier family 12 (sodium/chloride transporter), member 3 |
| chr3_+_3994248 | 0.19 |
|
|
|
| chr3_+_19911730 | 0.19 |
|
|
|
| chr22_-_10135735 | 0.19 |
ENSDART00000142012
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr5_-_31301280 | 0.19 |
ENSDART00000141446
|
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr21_+_11656401 | 0.18 |
ENSDART00000102404
|
ell2
|
elongation factor, RNA polymerase II, 2 |
| chr4_-_73485366 | 0.18 |
ENSDART00000122898
|
phf21b
|
PHD finger protein 21B |
| chr7_+_58718614 | 0.17 |
ENSDART00000157873
|
hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr7_-_5366526 | 0.17 |
|
|
|
| chr5_-_40894693 | 0.16 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr15_-_14616083 | 0.16 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr14_-_4038642 | 0.16 |
ENSDART00000077348
|
casp3b
|
caspase 3, apoptosis-related cysteine peptidase b |
| chr18_+_15964146 | 0.15 |
|
|
|
| chr15_-_36490729 | 0.15 |
|
|
|
| chr22_-_36561217 | 0.15 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr8_+_17479651 | 0.15 |
|
|
|
| chr10_-_20545884 | 0.15 |
|
|
|
| chr2_+_38390887 | 0.15 |
ENSDART00000113111
|
psmb5
|
proteasome subunit beta 5 |
| chr20_+_22900302 | 0.14 |
ENSDART00000131132
|
scfd2
|
sec1 family domain containing 2 |
| chr3_+_32400841 | 0.14 |
ENSDART00000150981
|
nosip
|
nitric oxide synthase interacting protein |
| chr24_-_14447136 | 0.14 |
|
|
|
| chr12_+_18777525 | 0.13 |
ENSDART00000105858
|
xrcc6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr18_-_1347210 | 0.13 |
ENSDART00000171402
|
pigb
|
phosphatidylinositol glycan anchor biosynthesis, class B |
| chr8_-_49739908 | 0.13 |
ENSDART00000138810
|
gkap1
|
G kinase anchoring protein 1 |
| chr19_+_46371192 | 0.12 |
ENSDART00000169710
|
utp23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr11_+_11284030 | 0.12 |
ENSDART00000026814
|
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr6_+_7376428 | 0.12 |
ENSDART00000057823
|
pa2g4a
|
proliferation-associated 2G4, a |
| chr13_-_33529333 | 0.12 |
ENSDART00000133073
|
rrbp1a
|
ribosome binding protein 1a |
| chr6_+_20402297 | 0.12 |
|
|
|
| chr4_-_2370333 | 0.12 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr22_-_16128314 | 0.12 |
ENSDART00000009464
|
slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr9_-_34691081 | 0.12 |
ENSDART00000114043
|
ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr4_-_62022037 | 0.12 |
|
|
|
| chr5_+_69013399 | 0.11 |
ENSDART00000162057
ENSDART00000166893 |
setd8b
|
SET domain containing (lysine methyltransferase) 8b |
| chr6_+_3556296 | 0.11 |
ENSDART00000041627
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr5_-_68712571 | 0.11 |
ENSDART00000109487
|
CABZ01032476.1
|
ENSDARG00000074645 |
| chr12_+_22438908 | 0.10 |
ENSDART00000172066
|
polr2a
|
polymerase (RNA) II (DNA directed) polypeptide A |
| chr4_-_73485324 | 0.10 |
ENSDART00000122898
|
phf21b
|
PHD finger protein 21B |
| KN150123v1_-_37086 | 0.10 |
|
|
|
| chr2_+_21267793 | 0.09 |
ENSDART00000099913
|
pla2g4aa
|
phospholipase A2, group IVAa (cytosolic, calcium-dependent) |
| chr22_-_12905648 | 0.09 |
ENSDART00000157820
|
mfsd6a
|
major facilitator superfamily domain containing 6a |
| chr23_+_29956616 | 0.09 |
ENSDART00000148706
|
aurkaip1
|
aurora kinase A interacting protein 1 |
| chr7_+_72030256 | 0.09 |
ENSDART00000172021
|
tollip
|
toll interacting protein |
| chr16_+_38251717 | 0.09 |
ENSDART00000044971
|
myo1eb
|
myosin IE, b |
| chr12_-_41128584 | 0.09 |
|
|
|
| chr19_-_48327666 | 0.09 |
|
|
|
| chr5_-_68712851 | 0.08 |
ENSDART00000109487
|
CABZ01032476.1
|
ENSDARG00000074645 |
| chr25_+_1046304 | 0.07 |
ENSDART00000172662
|
fam96a
|
family with sequence similarity 96, member A |
| chr8_+_31707960 | 0.07 |
ENSDART00000013434
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr23_-_29952057 | 0.07 |
ENSDART00000058407
|
slc25a33
|
solute carrier family 25 (pyrimidine nucleotide carrier), member 33 |
| chr7_+_8834138 | 0.07 |
ENSDART00000173250
|
pcsk6
|
proprotein convertase subtilisin/kexin type 6 |
| chr12_-_3042545 | 0.06 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr13_-_36265415 | 0.05 |
ENSDART00000133740
ENSDART00000100217 |
actn1
|
actinin, alpha 1 |
| chr5_+_31590746 | 0.05 |
|
|
|
| chr7_-_48392412 | 0.05 |
|
|
|
| KN149711v1_-_7827 | 0.05 |
ENSDART00000171410
|
ENSDARG00000104905
|
ENSDARG00000104905 |
| chr3_+_28808901 | 0.05 |
ENSDART00000141904
ENSDART00000077221 |
lgals1l1
|
lectin, galactoside-binding, soluble, 1 (galectin 1)-like 1 |
| chr12_+_24221087 | 0.04 |
ENSDART00000088178
|
nrxn1a
|
neurexin 1a |
| chr20_-_23272102 | 0.04 |
ENSDART00000167197
|
spata18
|
spermatogenesis associated 18 |
| chr1_-_50215233 | 0.04 |
ENSDART00000137648
|
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr13_+_35402382 | 0.04 |
ENSDART00000163368
|
wdr27
|
WD repeat domain 27 |
| chr2_-_55565099 | 0.03 |
ENSDART00000149062
|
rab8a
|
RAB8A, member RAS oncogene family |
| chr15_+_9351211 | 0.03 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr25_+_4729095 | 0.03 |
ENSDART00000163839
|
ap4e1
|
adaptor-related protein complex 4, epsilon 1 subunit |
| chr8_-_53593880 | 0.02 |
ENSDART00000015554
|
ubiad1
|
UbiA prenyltransferase domain containing 1 |
| chr14_-_6831278 | 0.02 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr2_+_11094785 | 0.02 |
ENSDART00000091497
|
tceanc2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| chr1_+_53384116 | 0.02 |
ENSDART00000149760
|
triobpa
|
TRIO and F-actin binding protein a |
| chr20_+_40560443 | 0.02 |
ENSDART00000153119
|
serinc1
|
serine incorporator 1 |
| chr6_-_16590883 | 0.02 |
ENSDART00000153552
|
nomo
|
nodal modulator |
| chr19_+_7505348 | 0.01 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr6_-_34854950 | 0.01 |
ENSDART00000169605
|
mier1a
|
mesoderm induction early response 1a, transcriptional regulator |
| chr7_-_57680717 | 0.01 |
|
|
|
| chr20_+_36098607 | 0.01 |
ENSDART00000021456
|
opn5
|
opsin 5 |
| chr19_+_46371377 | 0.00 |
ENSDART00000169710
|
utp23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0019344 | homoserine metabolic process(GO:0009092) cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) transsulfuration(GO:0019346) |
| 0.2 | 1.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 0.7 | GO:0046931 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.4 | GO:0002706 | regulation of lymphocyte mediated immunity(GO:0002706) |
| 0.1 | 0.3 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.1 | 1.0 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.1 | 0.3 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.1 | 0.4 | GO:0015817 | histidine transport(GO:0015817) |
| 0.1 | 0.3 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.1 | 0.2 | GO:0045813 | positive regulation of Wnt signaling pathway, calcium modulating pathway(GO:0045813) |
| 0.1 | 0.4 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 0.7 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 1.4 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 0.3 | GO:1903428 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 0.0 | 0.9 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.9 | GO:1990399 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.0 | 0.3 | GO:0019856 | pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.1 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.3 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.1 | GO:0022602 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) inflammatory response to wounding(GO:0090594) |
| 0.0 | 0.2 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.3 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.5 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:1902224 | cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.0 | 0.1 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.4 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 0.0 | 0.0 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.2 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 0.2 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.0 | 0.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.4 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.9 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.7 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 1.4 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.9 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0008665 | tRNA 2'-phosphotransferase activity(GO:0000215) 2'-phosphotransferase activity(GO:0008665) |
| 0.2 | 1.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.4 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.4 | GO:0015182 | L-histidine transmembrane transporter activity(GO:0005290) L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.1 | 0.5 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.1 | 0.4 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.1 | 0.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 1.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.2 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.6 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.4 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.5 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.2 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.9 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.2 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.0 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 0.7 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 0.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.9 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.7 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.4 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.3 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.6 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |