Project
DANIO-CODE
Navigation
Downloads
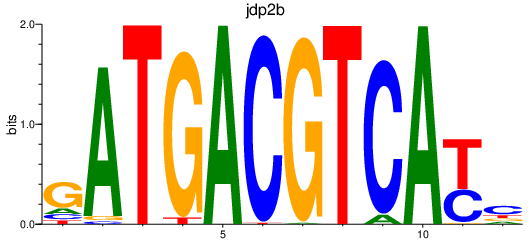
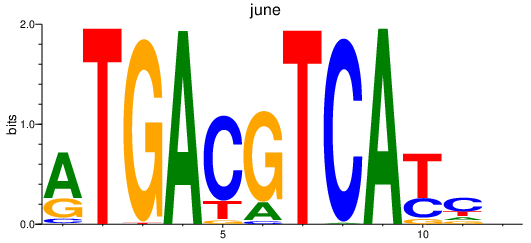
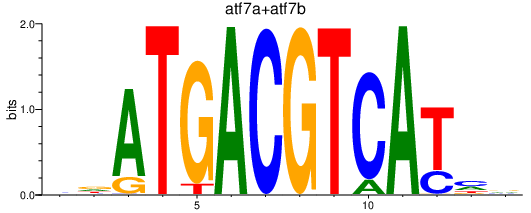
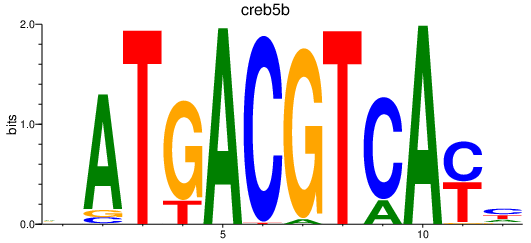
Results for jdp2b_june_atf7a+atf7b_creb5b
Z-value: 0.66
Transcription factors associated with jdp2b_june_atf7a+atf7b_creb5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
jdp2b
|
ENSDARG00000020133 | Jun dimerization protein 2b |
|
june
|
ENSDARG00000104090 | JunE proto-oncogene, AP-1 transcription factor subunit |
|
atf7a
|
ENSDARG00000011298 | activating transcription factor 7a |
|
atf7b
|
ENSDARG00000055481 | activating transcription factor 7b |
|
creb5b
|
ENSDARG00000070536 | cAMP responsive element binding protein 5b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| creb5b | dr10_dc_chr16_-_20901879_20901934 | 0.70 | 2.8e-03 | Click! |
| atf7a | dr10_dc_chr23_-_27569482_27569508 | 0.60 | 1.5e-02 | Click! |
| jdp2b | dr10_dc_chr20_+_46683031_46683038 | 0.48 | 6.2e-02 | Click! |
| atf7b | dr10_dc_chr6_-_39633603_39633697 | -0.45 | 8.0e-02 | Click! |
Activity profile of jdp2b_june_atf7a+atf7b_creb5b motif
Sorted Z-values of jdp2b_june_atf7a+atf7b_creb5b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of jdp2b_june_atf7a+atf7b_creb5b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_43611307 | 2.61 |
ENSDART00000151030
|
si:ch73-362m14.4
|
si:ch73-362m14.4 |
| chr15_+_28753020 | 2.55 |
ENSDART00000155815
|
nova2
|
neuro-oncological ventral antigen 2 |
| chr22_-_3747016 | 1.70 |
ENSDART00000083308
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr18_-_7184884 | 1.66 |
ENSDART00000135587
|
cd9a
|
CD9 molecule a |
| chr16_+_5284778 | 1.63 |
ENSDART00000156685
ENSDART00000156765 |
soga3a
soga3a
|
SOGA family member 3a SOGA family member 3a |
| chr5_-_23495973 | 1.53 |
ENSDART00000085969
|
map7d2a
|
MAP7 domain containing 2a |
| chr25_+_35179025 | 1.52 |
ENSDART00000149768
|
kif21a
|
kinesin family member 21A |
| chr20_+_29840979 | 1.51 |
ENSDART00000101603
|
kidins220b
|
kinase D-interacting substrate 220b |
| chr20_-_24223659 | 1.51 |
ENSDART00000153464
|
bach2b
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2b |
| KN149932v1_+_27584 | 1.48 |
|
|
|
| chr17_+_29328653 | 1.48 |
ENSDART00000086164
|
kctd3
|
potassium channel tetramerization domain containing 3 |
| chr19_-_7306228 | 1.41 |
ENSDART00000104799
|
col11a2
|
collagen, type XI, alpha 2 |
| chr2_-_30340646 | 1.41 |
ENSDART00000099078
|
jph1b
|
junctophilin 1b |
| chr7_-_14135850 | 1.40 |
ENSDART00000161442
|
CU467822.1
|
ENSDARG00000101584 |
| chr3_-_45420882 | 1.39 |
ENSDART00000161507
|
zgc:153426
|
zgc:153426 |
| chr5_+_55598181 | 1.38 |
ENSDART00000021159
|
lhx1b
|
LIM homeobox 1b |
| chr23_+_17856053 | 1.32 |
ENSDART00000154427
|
CR381647.1
|
ENSDARG00000097211 |
| chr23_-_7283514 | 1.28 |
ENSDART00000156369
|
CR589876.1
|
ENSDARG00000096997 |
| chr19_-_2011177 | 1.25 |
ENSDART00000108784
|
mturn
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr1_-_22170553 | 1.24 |
ENSDART00000139412
|
SMIM18
|
small integral membrane protein 18 |
| chr12_+_48643983 | 1.23 |
ENSDART00000168441
|
zgc:165653
|
zgc:165653 |
| chr24_+_4341242 | 1.23 |
ENSDART00000133360
|
ccny
|
cyclin Y |
| chr5_+_64175669 | 1.22 |
ENSDART00000050863
|
zgc:101858
|
zgc:101858 |
| chr19_-_7306136 | 1.21 |
ENSDART00000133179
|
col11a2
|
collagen, type XI, alpha 2 |
| chr2_-_10555152 | 1.19 |
ENSDART00000150166
|
gng12a
|
guanine nucleotide binding protein (G protein), gamma 12a |
| chr20_+_45531190 | 1.12 |
|
|
|
| chr8_+_933677 | 1.09 |
ENSDART00000149528
|
fabp1b.1
|
fatty acid binding protein 1b, tandem duplicate 1 |
| chr10_-_11806373 | 1.07 |
ENSDART00000009715
|
adamts6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
| chr18_-_7184699 | 1.06 |
ENSDART00000135587
|
cd9a
|
CD9 molecule a |
| chr8_+_15201828 | 1.02 |
ENSDART00000132216
|
paox1
|
polyamine oxidase (exo-N4-amino) 1 |
| chr13_+_17333810 | 0.99 |
ENSDART00000134181
|
CT025897.1
|
ENSDARG00000093577 |
| chr22_+_980836 | 0.98 |
ENSDART00000149743
|
pparda
|
peroxisome proliferator-activated receptor delta a |
| chr11_-_22211478 | 0.97 |
ENSDART00000065996
|
tmem183a
|
transmembrane protein 183A |
| chr23_-_5825236 | 0.96 |
ENSDART00000055087
|
phlda3
|
pleckstrin homology-like domain, family A, member 3 |
| chr21_+_25597814 | 0.94 |
ENSDART00000110705
|
tmem151a
|
transmembrane protein 151A |
| chr24_-_35897257 | 0.92 |
|
|
|
| chr21_-_23271127 | 0.92 |
ENSDART00000007806
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr9_+_54612545 | 0.92 |
ENSDART00000104475
|
tmsb4x
|
thymosin, beta 4 x |
| chr3_-_6078015 | 0.91 |
ENSDART00000165715
|
BX284638.1
|
ENSDARG00000098850 |
| chr23_+_17856104 | 0.90 |
ENSDART00000154427
|
CR381647.1
|
ENSDARG00000097211 |
| chr8_-_23102555 | 0.89 |
|
|
|
| chr9_-_296560 | 0.89 |
ENSDART00000160220
|
kif5aa
|
kinesin family member 5A, a |
| chr1_+_16681778 | 0.89 |
|
|
|
| KN150672v1_-_47685 | 0.87 |
|
|
|
| chr24_+_10276926 | 0.86 |
ENSDART00000145771
ENSDART00000157350 |
CT573344.1
|
ENSDARG00000093108 |
| chr19_-_39048324 | 0.86 |
ENSDART00000086717
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr12_-_25110150 | 0.83 |
ENSDART00000077188
|
cox7a3
|
cytochrome c oxidase subunit VIIa polypeptide 3 |
| chr13_-_40628582 | 0.82 |
ENSDART00000111641
ENSDART00000159255 |
morn4
|
MORN repeat containing 4 |
| chr2_+_54665352 | 0.80 |
|
|
|
| chr5_-_40894631 | 0.78 |
ENSDART00000121840
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr7_-_11812634 | 0.77 |
ENSDART00000101537
|
mex3b
|
mex-3 RNA binding family member B |
| chr9_-_296499 | 0.75 |
ENSDART00000165228
|
kif5aa
|
kinesin family member 5A, a |
| chr5_+_69036145 | 0.73 |
ENSDART00000174403
|
fsd1l
|
fibronectin type III and SPRY domain containing 1-like |
| chr16_-_55234426 | 0.73 |
ENSDART00000078887
|
tmem222a
|
transmembrane protein 222a |
| chr25_+_34340139 | 0.73 |
ENSDART00000061996
|
tmem231
|
transmembrane protein 231 |
| chr25_-_32344881 | 0.72 |
ENSDART00000012862
|
isl2a
|
ISL LIM homeobox 2a |
| chr3_-_6922811 | 0.70 |
|
|
|
| chr15_+_657554 | 0.69 |
ENSDART00000156007
|
si:ch73-144d13.8
|
si:ch73-144d13.8 |
| chr16_-_12282596 | 0.69 |
ENSDART00000110567
|
clstn3
|
calsyntenin 3 |
| chr22_+_20157574 | 0.68 |
|
|
|
| chr4_+_70746956 | 0.68 |
ENSDART00000172228
|
si:ch211-161m3.2
|
si:ch211-161m3.2 |
| chr21_+_22808694 | 0.67 |
ENSDART00000065555
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr9_-_13991784 | 0.65 |
ENSDART00000061156
|
raph1a
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1a |
| chr16_+_34569479 | 0.65 |
ENSDART00000041007
|
stmn1b
|
stathmin 1b |
| chr22_-_26971466 | 0.65 |
ENSDART00000087202
|
ENSDARG00000061256
|
ENSDARG00000061256 |
| chr18_-_5749619 | 0.63 |
ENSDART00000161538
|
cyp1a
|
cytochrome P450, family 1, subfamily A |
| chr8_+_21556157 | 0.63 |
ENSDART00000172190
|
ajap1
|
adherens junctions associated protein 1 |
| chr22_-_15567180 | 0.62 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr2_+_289138 | 0.62 |
ENSDART00000157246
|
znf1008
|
zinc finger protein 1008 |
| chr20_-_32543497 | 0.61 |
ENSDART00000026635
|
nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr21_-_45341242 | 0.60 |
ENSDART00000075438
|
cdkn2aipnl
|
CDKN2A interacting protein N-terminal like |
| chr8_+_8673307 | 0.59 |
ENSDART00000110854
|
elk1
|
ELK1, member of ETS oncogene family |
| chr24_-_7602759 | 0.59 |
ENSDART00000041714
|
atp6v0a1b
|
ATPase, H+ transporting, lysosomal V0 subunit a1b |
| chr3_-_3550073 | 0.57 |
ENSDART00000166667
|
tmem184bb
|
transmembrane protein 184bb |
| chr12_-_48643687 | 0.57 |
ENSDART00000178135
ENSDART00000176040 |
CABZ01078412.1
|
ENSDARG00000107260 |
| chr8_+_29733109 | 0.57 |
ENSDART00000020621
|
mapk4
|
mitogen-activated protein kinase 4 |
| chr13_-_40628537 | 0.56 |
ENSDART00000111641
ENSDART00000159255 |
morn4
|
MORN repeat containing 4 |
| chr24_+_9163779 | 0.56 |
|
|
|
| chr15_-_630492 | 0.56 |
ENSDART00000153884
|
si:ch73-144d13.5
|
si:ch73-144d13.5 |
| chr14_-_49951676 | 0.55 |
ENSDART00000021736
|
scocb
|
short coiled-coil protein b |
| chr5_+_44722544 | 0.54 |
ENSDART00000084411
|
adamts3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3 |
| chr19_-_19931617 | 0.54 |
ENSDART00000160582
|
creb5a
|
cAMP responsive element binding protein 5a |
| chr7_+_39957385 | 0.54 |
ENSDART00000052222
|
ptprn2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr3_-_16263697 | 0.53 |
ENSDART00000130575
ENSDART00000136465 |
eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr13_+_51663349 | 0.53 |
|
|
|
| chr18_-_19114558 | 0.53 |
ENSDART00000177621
|
dennd4a
|
DENN/MADD domain containing 4A |
| chr23_+_24141576 | 0.52 |
|
|
|
| chr23_+_25782195 | 0.52 |
ENSDART00000060059
|
rbms2b
|
RNA binding motif, single stranded interacting protein 2b |
| chr1_+_6862901 | 0.52 |
ENSDART00000015732
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr19_-_22184035 | 0.51 |
|
|
|
| chr10_-_43289221 | 0.51 |
ENSDART00000148293
|
tmem167a
|
transmembrane protein 167A |
| chr4_+_71392976 | 0.50 |
ENSDART00000167082
|
zgc:152938
|
zgc:152938 |
| chr5_+_5186074 | 0.50 |
|
|
|
| chr11_-_19414624 | 0.50 |
ENSDART00000157847
|
atxn7
|
ataxin 7 |
| chr12_-_36565562 | 0.50 |
ENSDART00000153259
|
si:ch211-216b21.2
|
si:ch211-216b21.2 |
| chr23_+_34891575 | 0.49 |
ENSDART00000013449
|
CHST13
|
carbohydrate sulfotransferase 13 |
| KN150072v1_+_1751 | 0.49 |
ENSDART00000168431
|
CABZ01033180.1
|
ENSDARG00000102665 |
| chr3_-_39208943 | 0.48 |
|
|
|
| chr24_+_24309660 | 0.48 |
|
|
|
| chr5_-_47519273 | 0.48 |
|
|
|
| chr15_+_1832874 | 0.48 |
ENSDART00000126253
|
fam124b
|
family with sequence similarity 124B |
| chr4_-_73559895 | 0.48 |
ENSDART00000162529
|
zgc:172139
|
zgc:172139 |
| chr14_+_16508093 | 0.48 |
ENSDART00000161201
|
limch1b
|
LIM and calponin homology domains 1b |
| chr1_-_50831155 | 0.47 |
ENSDART00000152719
|
spred2a
|
sprouty-related, EVH1 domain containing 2a |
| chr4_-_6365088 | 0.47 |
ENSDART00000140100
|
MDFIC
|
MyoD family inhibitor domain containing |
| chr3_+_43010408 | 0.47 |
ENSDART00000169061
|
CU138533.1
|
ENSDARG00000099842 |
| chr20_-_37733350 | 0.47 |
|
|
|
| chr15_-_1070691 | 0.46 |
ENSDART00000153569
|
si:dkey-77f5.4
|
si:dkey-77f5.4 |
| chr4_+_22754618 | 0.46 |
|
|
|
| chr19_+_24340673 | 0.46 |
|
|
|
| chr25_-_13057808 | 0.46 |
ENSDART00000172571
|
smpd3
|
sphingomyelin phosphodiesterase 3, neutral |
| chr7_-_34656224 | 0.46 |
ENSDART00000073397
|
nfatc3a
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3a |
| chr14_-_49115338 | 0.46 |
ENSDART00000056712
|
etfdh
|
electron-transferring-flavoprotein dehydrogenase |
| chr4_+_72559555 | 0.45 |
|
|
|
| chr23_-_624534 | 0.45 |
ENSDART00000132175
|
nadl1.1
|
neural adhesion molecule L1.1 |
| chr7_+_16348702 | 0.45 |
|
|
|
| chr24_+_192889 | 0.45 |
ENSDART00000067764
|
stk17a
|
serine/threonine kinase 17a |
| chr11_+_42203807 | 0.45 |
ENSDART00000067604
|
arf4a
|
ADP-ribosylation factor 4a |
| chr2_-_57133471 | 0.45 |
|
|
|
| chr13_+_24271932 | 0.44 |
ENSDART00000043002
|
rab1ab
|
RAB1A, member RAS oncogene family b |
| chr17_-_5453357 | 0.44 |
ENSDART00000004043
|
enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr11_+_3186396 | 0.44 |
ENSDART00000159459
|
pmela
|
premelanosome protein a |
| chr5_+_32282547 | 0.44 |
ENSDART00000135459
|
rpl12
|
ribosomal protein L12 |
| chr16_-_20901879 | 0.41 |
ENSDART00000103630
|
creb5b
|
cAMP responsive element binding protein 5b |
| chr9_+_219478 | 0.41 |
ENSDART00000164048
ENSDART00000161484 ENSDART00000171623 |
map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr13_-_49153890 | 0.40 |
ENSDART00000136991
|
irf2bp2a
|
interferon regulatory factor 2 binding protein 2a |
| chr9_-_306569 | 0.40 |
ENSDART00000166059
|
si:ch211-166e11.5
|
si:ch211-166e11.5 |
| chr7_-_12715948 | 0.40 |
ENSDART00000173115
|
rplp2l
|
ribosomal protein, large P2, like |
| chr4_-_25282256 | 0.40 |
ENSDART00000066936
|
tmem110l
|
transmembrane protein 110, like |
| chr9_-_6524104 | 0.40 |
ENSDART00000158468
|
nck2a
|
NCK adaptor protein 2a |
| chr5_-_40894693 | 0.39 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr4_-_38423096 | 0.39 |
|
|
|
| chr13_-_31339870 | 0.39 |
ENSDART00000076574
|
rtn1a
|
reticulon 1a |
| chr7_-_73866202 | 0.39 |
|
|
|
| chr5_+_64175494 | 0.39 |
ENSDART00000050863
|
zgc:101858
|
zgc:101858 |
| chr20_-_34898276 | 0.39 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr13_+_28574593 | 0.38 |
ENSDART00000126845
|
ldb1a
|
LIM domain binding 1a |
| chr17_-_47893203 | 0.38 |
|
|
|
| chr25_+_7147666 | 0.38 |
ENSDART00000104712
|
hmg20a
|
high mobility group 20A |
| chr4_-_71335661 | 0.37 |
|
|
|
| chr7_+_20377678 | 0.37 |
ENSDART00000173710
|
si:dkey-19b23.15
|
si:dkey-19b23.15 |
| chr9_+_31469848 | 0.37 |
|
|
|
| chr17_+_22936598 | 0.37 |
ENSDART00000155145
|
ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr11_+_44460956 | 0.37 |
ENSDART00000168531
|
irf2bp2b
|
interferon regulatory factor 2 binding protein 2b |
| chr6_+_15018104 | 0.37 |
ENSDART00000063648
|
nck2b
|
NCK adaptor protein 2b |
| chr23_+_19827551 | 0.37 |
ENSDART00000073442
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr23_+_29431173 | 0.36 |
ENSDART00000027255
|
tardbpl
|
TAR DNA binding protein, like |
| chr25_-_14573221 | 0.36 |
|
|
|
| chr5_-_31763197 | 0.36 |
ENSDART00000098045
|
gas1b
|
growth arrest-specific 1b |
| chr17_-_26893412 | 0.36 |
ENSDART00000045842
|
rcan3
|
regulator of calcineurin 3 |
| chr2_+_48448974 | 0.36 |
ENSDART00000023040
|
hes6
|
hes family bHLH transcription factor 6 |
| chr4_-_28651054 | 0.36 |
ENSDART00000167212
|
CABZ01050663.1
|
ENSDARG00000104584 |
| chr22_+_2606049 | 0.35 |
|
|
|
| chr10_-_9256025 | 0.35 |
ENSDART00000140830
|
arl15b
|
ADP-ribosylation factor-like 15b |
| chr17_+_1610578 | 0.35 |
ENSDART00000082101
|
ppp2r5ca
|
protein phosphatase 2, regulatory subunit B', gamma a |
| chr10_-_40905193 | 0.35 |
ENSDART00000172089
|
pcna
|
proliferating cell nuclear antigen |
| chr22_+_12327849 | 0.34 |
ENSDART00000178678
|
FO704572.1
|
ENSDARG00000106890 |
| chr10_+_28420109 | 0.34 |
ENSDART00000142016
|
ptrh2
|
peptidyl-tRNA hydrolase 2 |
| chr3_+_33168814 | 0.33 |
ENSDART00000114023
|
hspb9
|
heat shock protein, alpha-crystallin-related, 9 |
| chr4_-_72183874 | 0.33 |
ENSDART00000144382
|
si:dkey-262g12.12
|
si:dkey-262g12.12 |
| chr4_-_71322276 | 0.33 |
|
|
|
| chr6_+_27156169 | 0.33 |
ENSDART00000088364
|
kif1aa
|
kinesin family member 1Aa |
| chr8_-_410733 | 0.33 |
ENSDART00000151255
|
trim36
|
tripartite motif containing 36 |
| chr2_-_54965656 | 0.33 |
|
|
|
| chr4_-_1989775 | 0.33 |
ENSDART00000003790
|
pwp1
|
PWP1 homolog (S. cerevisiae) |
| chr9_+_48521741 | 0.33 |
ENSDART00000145972
|
ccdc173
|
coiled-coil domain containing 173 |
| chr17_-_5453436 | 0.33 |
ENSDART00000004043
|
enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr3_-_36130248 | 0.33 |
ENSDART00000126588
|
rac3a
|
ras-related C3 botulinum toxin substrate 3a (rho family, small GTP binding protein Rac3) |
| chr6_+_7092350 | 0.33 |
ENSDART00000049695
ENSDART00000136088 ENSDART00000083424 ENSDART00000125912 |
dzip1
|
DAZ interacting zinc finger protein 1 |
| chr2_+_23038657 | 0.33 |
ENSDART00000089012
|
kif1ab
|
kinesin family member 1Ab |
| chr23_-_45356039 | 0.33 |
ENSDART00000067630
|
ENSDARG00000039901
|
ENSDARG00000039901 |
| chr11_-_43860836 | 0.32 |
ENSDART00000047126
|
clcn4
|
chloride channel, voltage-sensitive 4 |
| chr5_+_2384929 | 0.32 |
|
|
|
| chr18_+_12089333 | 0.32 |
ENSDART00000112671
|
bicd1a
|
bicaudal D homolog 1a |
| chr25_+_16849382 | 0.32 |
ENSDART00000016591
|
fgf6a
|
fibroblast growth factor 6a |
| chr20_-_9474672 | 0.32 |
ENSDART00000152674
|
ENSDARG00000033201
|
ENSDARG00000033201 |
| chr4_-_25282161 | 0.31 |
ENSDART00000066936
|
tmem110l
|
transmembrane protein 110, like |
| chr14_-_24463906 | 0.31 |
ENSDART00000126199
|
slit3
|
slit homolog 3 (Drosophila) |
| chr4_+_12980657 | 0.31 |
|
|
|
| chr13_+_371940 | 0.31 |
ENSDART00000013007
|
degs1
|
delta(4)-desaturase, sphingolipid 1 |
| chr7_+_39934376 | 0.31 |
ENSDART00000173742
|
ncapg2
|
non-SMC condensin II complex, subunit G2 |
| chr20_-_54742983 | 0.31 |
|
|
|
| chr16_-_32695903 | 0.31 |
ENSDART00000136161
|
faxcb
|
failed axon connections homolog b |
| chr3_-_6922655 | 0.31 |
|
|
|
| chr19_-_39048402 | 0.30 |
ENSDART00000086717
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr17_+_12544451 | 0.30 |
ENSDART00000064509
ENSDART00000136830 |
stmn4l
|
stathmin-like 4, like |
| chr5_+_29489543 | 0.30 |
ENSDART00000078114
|
timm8b
|
translocase of inner mitochondrial membrane 8 homolog B (yeast) |
| chr14_+_905613 | 0.30 |
ENSDART00000111141
|
myoz3a
|
myozenin 3a |
| chr16_-_9978112 | 0.30 |
ENSDART00000149312
|
ncalda
|
neurocalcin delta a |
| chr4_-_71367238 | 0.29 |
ENSDART00000165327
|
CABZ01021450.2
|
ENSDARG00000100390 |
| chr23_-_4979524 | 0.29 |
ENSDART00000060714
|
atp6ap1a
|
ATPase, H+ transporting, lysosomal accessory protein 1a |
| chr5_+_71360122 | 0.29 |
|
|
|
| chr5_+_43365449 | 0.29 |
ENSDART00000113502
|
TMEM8B
|
si:dkey-84j12.1 |
| chr9_+_7570199 | 0.29 |
ENSDART00000081543
|
ptprna
|
protein tyrosine phosphatase, receptor type, Na |
| chr21_-_36886505 | 0.29 |
ENSDART00000113678
|
wwc1
|
WW and C2 domain containing 1 |
| chr25_+_19103272 | 0.29 |
ENSDART00000067323
|
hapln3
|
hyaluronan and proteoglycan link protein 3 |
| chr4_+_119964 | 0.29 |
ENSDART00000177177
ENSDART00000159996 |
tbk1
|
TANK-binding kinase 1 |
| chr4_-_1564624 | 0.29 |
ENSDART00000168633
|
BICD1 (1 of many)
|
BICD cargo adaptor 1 |
| chr24_+_37752791 | 0.29 |
ENSDART00000158181
|
wdr24
|
WD repeat domain 24 |
| chr3_+_31802145 | 0.28 |
ENSDART00000139644
|
lin7b
|
lin-7 homolog B (C. elegans) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.3 | 1.3 | GO:0030219 | megakaryocyte differentiation(GO:0030219) positive regulation of myeloid cell differentiation(GO:0045639) |
| 0.2 | 1.7 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) synaptic membrane adhesion(GO:0099560) |
| 0.2 | 1.0 | GO:0030194 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.2 | 0.9 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.2 | 2.6 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.2 | 1.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.2 | 1.2 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.2 | 1.2 | GO:0010888 | negative regulation of cholesterol storage(GO:0010887) negative regulation of lipid storage(GO:0010888) |
| 0.2 | 0.7 | GO:0010939 | regulation of necrotic cell death(GO:0010939) regulation of necroptotic process(GO:0060544) |
| 0.2 | 0.5 | GO:2001017 | regulation of retrograde axon cargo transport(GO:2001017) |
| 0.1 | 0.4 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 0.7 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.1 | 0.8 | GO:0051965 | regulation of synapse assembly(GO:0051963) positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.3 | GO:0014005 | microglia development(GO:0014005) |
| 0.1 | 0.5 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 2.6 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.5 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 0.3 | GO:0032475 | otolith formation(GO:0032475) |
| 0.1 | 0.6 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.4 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.4 | GO:0002762 | negative regulation of myeloid leukocyte differentiation(GO:0002762) |
| 0.1 | 0.3 | GO:0098971 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) axo-dendritic protein transport(GO:0099640) |
| 0.1 | 0.2 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 0.8 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.1 | 0.2 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 0.2 | GO:0019677 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.1 | 0.2 | GO:0072578 | neurotransmitter-gated ion channel clustering(GO:0072578) |
| 0.1 | 0.2 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) |
| 0.1 | 0.8 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 2.6 | GO:0007338 | single fertilization(GO:0007338) |
| 0.1 | 0.5 | GO:0007045 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.1 | 0.3 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.2 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.1 | 0.1 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.2 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.3 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.5 | GO:0021681 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.3 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.1 | GO:0003321 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.0 | 0.3 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 1.3 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.2 | GO:1901739 | regulation of myoblast fusion(GO:1901739) |
| 0.0 | 0.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 3.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.2 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.4 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0050864 | regulation of B cell activation(GO:0050864) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 1.6 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.2 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0061323 | cell proliferation involved in heart morphogenesis(GO:0061323) regulation of cell proliferation involved in heart morphogenesis(GO:2000136) |
| 0.0 | 0.4 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.6 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 2.1 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.2 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.1 | GO:0043247 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.0 | 0.2 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.0 | 0.2 | GO:0036302 | atrioventricular canal development(GO:0036302) |
| 0.0 | 2.8 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.2 | GO:0015671 | response to activity(GO:0014823) oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 1.0 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.3 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.1 | GO:0033986 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.0 | 0.0 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.0 | GO:0051299 | mitotic centrosome separation(GO:0007100) centrosome separation(GO:0051299) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.3 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.4 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 1.9 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.4 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.2 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 1.1 | GO:0033993 | response to lipid(GO:0033993) |
| 0.0 | 0.6 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.4 | GO:0030834 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) negative regulation of protein complex disassembly(GO:0043242) actin filament capping(GO:0051693) regulation of protein depolymerization(GO:1901879) negative regulation of protein depolymerization(GO:1901880) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 2.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.5 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 1.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 0.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 3.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.6 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 2.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 0.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 1.5 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 1.3 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 3.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.2 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.9 | GO:0005884 | actin filament(GO:0005884) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.3 | 2.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.3 | 0.8 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.3 | 0.8 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.2 | 0.6 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.2 | 2.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.4 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.5 | GO:0032034 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.1 | 0.3 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.1 | 1.2 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.5 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 1.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 1.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.4 | GO:0019808 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.1 | 0.3 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.4 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 1.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 0.5 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 0.3 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.1 | 0.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 3.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.5 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.3 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 1.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.4 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.3 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 1.3 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 1.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 3.3 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.5 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.2 | GO:0019825 | oxygen transporter activity(GO:0005344) oxygen binding(GO:0019825) |
| 0.0 | 0.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.1 | GO:0004935 | adrenergic receptor activity(GO:0004935) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.2 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 0.7 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.5 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.5 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 1.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.9 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 0.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 0.6 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |