Project
DANIO-CODE
Navigation
Downloads
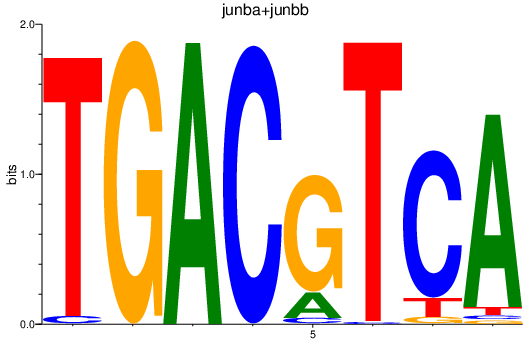
Results for junba+junbb
Z-value: 0.78
Transcription factors associated with junba+junbb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
junba
|
ENSDARG00000074378 | JunB proto-oncogene, AP-1 transcription factor subunit a |
|
junbb
|
ENSDARG00000104773 | JunB proto-oncogene, AP-1 transcription factor subunit b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| junba | dr10_dc_chr1_-_51100156_51100163 | 0.09 | 7.5e-01 | Click! |
Activity profile of junba+junbb motif
Sorted Z-values of junba+junbb motif
Network of associatons between targets according to the STRING database.
First level regulatory network of junba+junbb
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_49749441 | 1.06 |
ENSDART00000136165
|
lyst
|
lysosomal trafficking regulator |
| chr1_+_6862901 | 0.97 |
ENSDART00000015732
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr9_+_219478 | 0.95 |
ENSDART00000164048
ENSDART00000161484 ENSDART00000171623 |
map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr9_-_39195468 | 0.93 |
ENSDART00000014207
|
myl1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr17_+_1610578 | 0.92 |
ENSDART00000082101
|
ppp2r5ca
|
protein phosphatase 2, regulatory subunit B', gamma a |
| chr3_+_33168814 | 0.78 |
ENSDART00000114023
|
hspb9
|
heat shock protein, alpha-crystallin-related, 9 |
| chr23_+_44817648 | 0.66 |
ENSDART00000143688
|
dlg4b
|
discs, large homolog 4b (Drosophila) |
| chr23_-_7283514 | 0.62 |
ENSDART00000156369
|
CR589876.1
|
ENSDARG00000096997 |
| chr20_+_35010140 | 0.61 |
ENSDART00000172001
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr13_-_49749469 | 0.61 |
ENSDART00000136165
|
lyst
|
lysosomal trafficking regulator |
| chr17_+_25163592 | 0.59 |
ENSDART00000100277
|
cln8
|
CLN8, transmembrane ER and ERGIC protein |
| chr22_+_18228143 | 0.59 |
ENSDART00000141535
|
BX664610.1
|
ENSDARG00000095557 |
| chr13_-_47266563 | 0.58 |
ENSDART00000131239
|
acoxl
|
acyl-CoA oxidase-like |
| chr9_+_4188263 | 0.57 |
ENSDART00000164543
|
kalrna
|
kalirin RhoGEF kinase a |
| chr6_+_10098305 | 0.57 |
ENSDART00000151477
|
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr17_-_12231165 | 0.57 |
ENSDART00000080927
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr22_-_38635149 | 0.56 |
ENSDART00000098330
|
nppc
|
natriuretic peptide C |
| chr25_-_22955066 | 0.56 |
ENSDART00000024633
|
dusp8a
|
dual specificity phosphatase 8a |
| chr2_+_23038657 | 0.55 |
ENSDART00000089012
|
kif1ab
|
kinesin family member 1Ab |
| chr11_-_18074946 | 0.53 |
|
|
|
| chr11_+_3186396 | 0.52 |
ENSDART00000159459
|
pmela
|
premelanosome protein a |
| chr1_-_22170553 | 0.51 |
ENSDART00000139412
|
SMIM18
|
small integral membrane protein 18 |
| chr8_-_14113377 | 0.50 |
ENSDART00000090427
|
si:dkey-6n6.1
|
si:dkey-6n6.1 |
| chr2_-_42566339 | 0.50 |
ENSDART00000121452
|
slco5a1b
|
solute carrier organic anion transporter family member 5A1b |
| chr13_+_15685540 | 0.50 |
ENSDART00000137061
|
klc1a
|
kinesin light chain 1a |
| chr2_+_10023967 | 0.49 |
ENSDART00000148227
|
anxa13l
|
annexin A13, like |
| chr5_+_53854279 | 0.49 |
ENSDART00000165889
|
tprn
|
taperin |
| chr12_-_308929 | 0.49 |
ENSDART00000066579
ENSDART00000171396 |
pts
|
6-pyruvoyltetrahydropterin synthase |
| chr17_+_25168837 | 0.49 |
ENSDART00000148431
|
cln8
|
CLN8, transmembrane ER and ERGIC protein |
| chr3_+_32272097 | 0.48 |
ENSDART00000156982
|
si:ch211-195b15.8
|
si:ch211-195b15.8 |
| chr25_-_36512943 | 0.48 |
ENSDART00000114508
|
hprt1l
|
hypoxanthine phosphoribosyltransferase 1, like |
| chr5_+_8542777 | 0.47 |
ENSDART00000091463
|
idua
|
iduronidase, alpha-L- |
| chr1_+_31131899 | 0.47 |
ENSDART00000075260
|
inab
|
internexin neuronal intermediate filament protein, alpha b |
| chr21_-_43611307 | 0.46 |
ENSDART00000151030
|
si:ch73-362m14.4
|
si:ch73-362m14.4 |
| chr18_+_8959686 | 0.45 |
ENSDART00000145226
|
si:ch211-233h19.2
|
si:ch211-233h19.2 |
| chr7_-_8635687 | 0.43 |
ENSDART00000081620
|
vax2
|
ventral anterior homeobox 2 |
| chr17_+_50577102 | 0.42 |
ENSDART00000127374
|
ddhd1a
|
DDHD domain containing 1a |
| chr12_-_17585587 | 0.41 |
ENSDART00000142427
|
pvalb3
|
parvalbumin 3 |
| chr13_-_24248675 | 0.41 |
ENSDART00000046360
|
rhoua
|
ras homolog family member Ua |
| chr5_-_23495973 | 0.41 |
ENSDART00000085969
|
map7d2a
|
MAP7 domain containing 2a |
| chr19_+_16318256 | 0.40 |
ENSDART00000137189
|
ptprua
|
protein tyrosine phosphatase, receptor type, U, a |
| chr14_+_23631214 | 0.40 |
ENSDART00000172149
|
sh3rf2
|
SH3 domain containing ring finger 2 |
| chr19_+_46380082 | 0.40 |
ENSDART00000158781
|
si:ch211-153f2.7
|
si:ch211-153f2.7 |
| chr5_-_47519273 | 0.39 |
|
|
|
| chr23_-_7889744 | 0.39 |
ENSDART00000164117
|
myt1b
|
myelin transcription factor 1b |
| chr5_+_32314200 | 0.39 |
ENSDART00000051336
|
cyr61l1
|
cysteine-rich, angiogenic inducer 61 like 1 |
| chr3_+_43010408 | 0.38 |
ENSDART00000169061
|
CU138533.1
|
ENSDARG00000099842 |
| chr23_+_28656263 | 0.38 |
ENSDART00000020296
|
nadl1.2
|
neural adhesion molecule L1.2 |
| chr23_-_624534 | 0.38 |
ENSDART00000132175
|
nadl1.1
|
neural adhesion molecule L1.1 |
| chr15_+_36197093 | 0.38 |
ENSDART00000156697
|
col4a3
|
collagen, type IV, alpha 3 |
| chr17_+_12544451 | 0.37 |
ENSDART00000064509
ENSDART00000136830 |
stmn4l
|
stathmin-like 4, like |
| chr19_-_10635235 | 0.37 |
ENSDART00000104539
|
lim2.4
|
lens intrinsic membrane protein 2.4 |
| chr5_+_19802818 | 0.37 |
|
|
|
| chr3_-_39208943 | 0.37 |
|
|
|
| chr14_+_905613 | 0.36 |
ENSDART00000111141
|
myoz3a
|
myozenin 3a |
| chr22_+_12327849 | 0.36 |
ENSDART00000178678
|
FO704572.1
|
ENSDARG00000106890 |
| chr20_-_52133924 | 0.36 |
ENSDART00000074325
|
dusp10
|
dual specificity phosphatase 10 |
| chr14_-_41101660 | 0.35 |
ENSDART00000003170
|
mid1ip1l
|
MID1 interacting protein 1, like |
| chr7_-_51474531 | 0.35 |
ENSDART00000083190
|
hdac8
|
histone deacetylase 8 |
| chr8_-_9081109 | 0.34 |
ENSDART00000176850
|
slc6a8
|
solute carrier family 6 (neurotransmitter transporter), member 8 |
| chr7_-_57727148 | 0.34 |
|
|
|
| chr18_+_10815994 | 0.34 |
ENSDART00000028938
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr23_-_30858769 | 0.34 |
ENSDART00000131285
|
myt1a
|
myelin transcription factor 1a |
| chr6_-_36817224 | 0.34 |
ENSDART00000160669
|
opa1
|
optic atrophy 1 (autosomal dominant) |
| chr6_+_27156169 | 0.34 |
ENSDART00000088364
|
kif1aa
|
kinesin family member 1Aa |
| chr11_-_7310284 | 0.34 |
ENSDART00000091664
|
apc2
|
adenomatosis polyposis coli 2 |
| chr18_+_10815958 | 0.34 |
ENSDART00000028938
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr20_+_3095763 | 0.33 |
ENSDART00000133435
|
CEP170B
|
centrosomal protein 170B |
| chr22_+_30381331 | 0.33 |
ENSDART00000104751
|
mxi1
|
max interactor 1, dimerization protein |
| chr9_-_55878647 | 0.33 |
|
|
|
| chr23_+_19827551 | 0.33 |
ENSDART00000073442
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr22_+_748767 | 0.32 |
ENSDART00000125347
|
cry1bb
|
cryptochrome circadian clock 1bb |
| chr5_+_71367929 | 0.32 |
ENSDART00000149910
|
abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr5_+_36250811 | 0.32 |
ENSDART00000051186
|
nccrp1
|
non-specific cytotoxic cell receptor protein 1 |
| chr6_+_33261144 | 0.32 |
ENSDART00000157428
|
pik3r3b
|
phosphoinositide-3-kinase, regulatory subunit 3b (gamma) |
| chr15_-_36196994 | 0.32 |
ENSDART00000163061
|
col4a4
|
collagen, type IV, alpha 4 |
| chr19_-_30816780 | 0.32 |
ENSDART00000103475
|
agr2
|
anterior gradient 2 |
| chr10_-_26782374 | 0.31 |
ENSDART00000162710
|
fgf13b
|
fibroblast growth factor 13b |
| chr18_+_1018614 | 0.31 |
ENSDART00000161206
|
pkma
|
pyruvate kinase, muscle, a |
| chr20_+_54762145 | 0.31 |
ENSDART00000166592
|
ENSDARG00000102965
|
ENSDARG00000102965 |
| chr17_-_52735250 | 0.31 |
|
|
|
| chr21_+_25017684 | 0.30 |
ENSDART00000065132
|
zgc:171740
|
zgc:171740 |
| chr22_+_38928192 | 0.30 |
ENSDART00000133067
ENSDART00000085701 |
AL953867.2
senp5
|
ENSDARG00000095689 SUMO1/sentrin specific peptidase 5 |
| chr5_+_26840147 | 0.30 |
ENSDART00000064701
|
loxl2b
|
lysyl oxidase-like 2b |
| chr12_-_10474942 | 0.30 |
ENSDART00000106163
ENSDART00000124562 |
zgc:152977
|
zgc:152977 |
| chr8_+_28046911 | 0.29 |
ENSDART00000078533
|
kcnd3
|
potassium voltage-gated channel, Shal-related subfamily, member 3 |
| chr7_+_23636549 | 0.29 |
ENSDART00000045479
|
syt4
|
synaptotagmin IV |
| chr10_+_32702525 | 0.29 |
ENSDART00000137244
|
zbtb21
|
zinc finger and BTB domain containing 21 |
| chr1_-_50962689 | 0.29 |
ENSDART00000130828
|
cnrip1a
|
cannabinoid receptor interacting protein 1a |
| chr1_-_24297477 | 0.28 |
|
|
|
| chr7_-_21978889 | 0.28 |
|
|
|
| chr3_+_14314116 | 0.28 |
ENSDART00000170927
|
cnn1b
|
calponin 1, basic, smooth muscle, b |
| chr8_+_51721225 | 0.28 |
ENSDART00000129372
|
klhl22
|
kelch-like family member 22 |
| chr12_-_18981467 | 0.28 |
ENSDART00000060561
|
csdc2a
|
cold shock domain containing C2, RNA binding a |
| chr12_-_17079132 | 0.28 |
ENSDART00000020541
|
lipf
|
lipase, gastric |
| chr1_-_54269904 | 0.28 |
ENSDART00000129710
|
crtac1a
|
cartilage acidic protein 1a |
| chr15_-_28267088 | 0.27 |
ENSDART00000004200
|
sarm1
|
sterile alpha and TIR motif containing 1 |
| chr8_-_14446883 | 0.27 |
ENSDART00000057644
|
lhx4
|
LIM homeobox 4 |
| chr6_+_27676752 | 0.27 |
ENSDART00000159624
ENSDART00000049177 |
rab6ba
|
RAB6B, member RAS oncogene family a |
| chr13_-_22943797 | 0.27 |
ENSDART00000143210
|
srgn
|
serglycin |
| chr10_+_5688293 | 0.27 |
ENSDART00000172632
|
pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr2_+_35505943 | 0.27 |
ENSDART00000131248
|
CR855337.1
|
ENSDARG00000094295 |
| chr6_-_18891908 | 0.27 |
ENSDART00000074327
|
igfbp2a
|
insulin-like growth factor binding protein 2a |
| chr3_-_22083446 | 0.26 |
ENSDART00000154818
|
maptb
|
microtubule-associated protein tau b |
| chr8_-_27839244 | 0.26 |
ENSDART00000136562
|
cttnbp2nlb
|
CTTNBP2 N-terminal like b |
| chr4_-_16356071 | 0.26 |
ENSDART00000079521
|
kera
|
keratocan |
| chr20_-_34898276 | 0.26 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr11_+_24826881 | 0.26 |
ENSDART00000157435
|
ndrg3a
|
ndrg family member 3a |
| chr23_-_624480 | 0.26 |
ENSDART00000132175
|
nadl1.1
|
neural adhesion molecule L1.1 |
| chr7_-_8635989 | 0.26 |
ENSDART00000081620
|
vax2
|
ventral anterior homeobox 2 |
| chr24_-_3751435 | 0.26 |
ENSDART00000139596
|
adarb2
|
adenosine deaminase, RNA-specific, B2 (non-functional) |
| chr17_-_22552999 | 0.26 |
ENSDART00000141523
|
exo1
|
exonuclease 1 |
| chr19_-_39048324 | 0.26 |
ENSDART00000086717
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr23_+_12225922 | 0.26 |
ENSDART00000136046
|
ppp1r3da
|
protein phosphatase 1, regulatory subunit 3Da |
| chr25_+_16849382 | 0.25 |
ENSDART00000016591
|
fgf6a
|
fibroblast growth factor 6a |
| chr8_-_38126693 | 0.25 |
ENSDART00000112331
|
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr9_+_21725007 | 0.25 |
ENSDART00000045093
|
arhgef7a
|
Rho guanine nucleotide exchange factor (GEF) 7a |
| chr22_+_26737422 | 0.25 |
|
|
|
| chr1_-_45941709 | 0.25 |
ENSDART00000053232
|
cdadc1
|
cytidine and dCMP deaminase domain containing 1 |
| chr2_-_43318809 | 0.25 |
ENSDART00000132588
|
crema
|
cAMP responsive element modulator a |
| KN150199v1_+_10711 | 0.25 |
|
|
|
| chr12_+_44829938 | 0.25 |
ENSDART00000098932
|
wbp2
|
WW domain binding protein 2 |
| chr4_-_12915292 | 0.24 |
ENSDART00000067135
|
msrb3
|
methionine sulfoxide reductase B3 |
| chr3_-_45420882 | 0.24 |
ENSDART00000161507
|
zgc:153426
|
zgc:153426 |
| chr12_+_32936230 | 0.24 |
ENSDART00000153146
|
rbfox3a
|
RNA binding protein, fox-1 homolog (C. elegans) 3a |
| chr18_-_6336677 | 0.24 |
ENSDART00000169844
|
iqsec3b
|
IQ motif and Sec7 domain 3b |
| chr21_-_4530904 | 0.24 |
ENSDART00000031425
|
zgc:55582
|
zgc:55582 |
| chr22_+_30097988 | 0.24 |
ENSDART00000040538
|
add3a
|
adducin 3 (gamma) a |
| chr22_+_24596299 | 0.24 |
ENSDART00000158303
ENSDART00000160924 |
mcoln2
|
mucolipin 2 |
| chr12_+_31523147 | 0.24 |
ENSDART00000153129
|
dnmbp
|
dynamin binding protein |
| chr25_-_8475902 | 0.24 |
ENSDART00000176751
|
CU929451.3
|
ENSDARG00000107886 |
| chr20_-_39200252 | 0.23 |
ENSDART00000037318
ENSDART00000143379 |
rcan2
|
regulator of calcineurin 2 |
| chr19_-_10476653 | 0.23 |
ENSDART00000109488
|
zgc:194578
|
zgc:194578 |
| chr2_-_16548451 | 0.23 |
ENSDART00000152031
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr20_-_37733350 | 0.23 |
|
|
|
| chr15_-_34560555 | 0.23 |
ENSDART00000147582
|
agmo
|
alkylglycerol monooxygenase |
| chr11_-_80746 | 0.23 |
|
|
|
| chr13_+_30033024 | 0.23 |
ENSDART00000040926
|
eif4ebp2
|
eukaryotic translation initiation factor 4E binding protein 2 |
| chr14_-_30739765 | 0.23 |
ENSDART00000173282
|
mbnl3
|
muscleblind-like splicing regulator 3 |
| chr20_-_18836593 | 0.23 |
ENSDART00000142837
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| KN150108v1_+_18276 | 0.23 |
|
|
|
| chr3_-_36130248 | 0.23 |
ENSDART00000126588
|
rac3a
|
ras-related C3 botulinum toxin substrate 3a (rho family, small GTP binding protein Rac3) |
| chr11_+_19894390 | 0.22 |
ENSDART00000103997
|
fezf2
|
FEZ family zinc finger 2 |
| chr7_-_37283894 | 0.22 |
ENSDART00000148905
ENSDART00000150229 |
cylda
|
cylindromatosis (turban tumor syndrome), a |
| chr13_-_49707003 | 0.22 |
|
|
|
| chr9_+_16437408 | 0.22 |
ENSDART00000006787
|
epha3
|
eph receptor A3 |
| chr12_+_19183576 | 0.22 |
ENSDART00000066391
|
csnk1e
|
casein kinase 1, epsilon |
| chr4_+_70728870 | 0.22 |
ENSDART00000122809
|
si:ch211-161m3.2
|
si:ch211-161m3.2 |
| chr9_+_31204464 | 0.22 |
|
|
|
| chr8_+_50202152 | 0.22 |
ENSDART00000099863
|
slc25a37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr23_+_19728953 | 0.22 |
ENSDART00000104441
|
abhd6b
|
abhydrolase domain containing 6b |
| chr9_+_21911860 | 0.22 |
ENSDART00000102021
|
sox1a
|
SRY (sex determining region Y)-box 1a |
| chr13_+_1484997 | 0.22 |
|
|
|
| chr22_+_18762028 | 0.22 |
ENSDART00000105399
|
cbarpb
|
calcium channel, voltage-dependent, beta subunit associated regulatory protein b |
| chr19_-_7069850 | 0.22 |
ENSDART00000145741
|
znf384l
|
zinc finger protein 384 like |
| chr25_-_36513113 | 0.21 |
ENSDART00000025494
|
hprt1l
|
hypoxanthine phosphoribosyltransferase 1, like |
| chr10_+_20435265 | 0.21 |
ENSDART00000160803
|
r3hcc1
|
R3H domain and coiled-coil containing 1 |
| chr18_+_16759954 | 0.21 |
ENSDART00000061265
|
rnf141
|
ring finger protein 141 |
| chr6_+_54857805 | 0.21 |
|
|
|
| chr8_+_50201986 | 0.21 |
ENSDART00000099863
|
slc25a37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr25_+_23494481 | 0.21 |
ENSDART00000089199
|
cpt1ab
|
carnitine palmitoyltransferase 1Ab (liver) |
| chr14_-_26999410 | 0.21 |
ENSDART00000159727
|
pcdh11
|
protocadherin 11 |
| chr21_+_28710341 | 0.21 |
ENSDART00000014058
|
zgc:100829
|
zgc:100829 |
| chr13_-_36420228 | 0.21 |
|
|
|
| chr2_+_26273986 | 0.21 |
ENSDART00000137746
|
slc7a14a
|
solute carrier family 7, member 14a |
| chr18_-_12483329 | 0.21 |
ENSDART00000092522
|
si:ch211-1e14.1
|
si:ch211-1e14.1 |
| chr10_+_1384178 | 0.21 |
ENSDART00000150096
|
gdnfa
|
glial cell derived neurotrophic factor a |
| chr23_+_17294623 | 0.21 |
ENSDART00000054761
|
nol4lb
|
nucleolar protein 4-like b |
| chr25_-_19889225 | 0.21 |
|
|
|
| chr6_-_36817334 | 0.21 |
ENSDART00000161928
|
opa1
|
optic atrophy 1 (autosomal dominant) |
| chr14_+_34146377 | 0.21 |
ENSDART00000131861
|
tmsb2
|
thymosin beta 2 |
| chr12_-_292556 | 0.21 |
ENSDART00000152608
|
CU611036.1
|
ENSDARG00000079888 |
| chr15_+_36196988 | 0.20 |
ENSDART00000156697
|
col4a3
|
collagen, type IV, alpha 3 |
| chr4_+_9477541 | 0.20 |
ENSDART00000030738
|
lmf2b
|
lipase maturation factor 2b |
| chr11_-_16067646 | 0.20 |
|
|
|
| chr10_+_32160464 | 0.20 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr9_-_41705830 | 0.20 |
ENSDART00000135821
|
mfsd6b
|
major facilitator superfamily domain containing 6b |
| chr18_-_21116872 | 0.20 |
ENSDART00000100791
|
si:dkey-12e7.1
|
si:dkey-12e7.1 |
| chr22_-_10574773 | 0.20 |
ENSDART00000091777
|
bap1
|
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) |
| chr12_+_14040873 | 0.20 |
ENSDART00000078033
|
ENSDARG00000055642
|
ENSDARG00000055642 |
| chr9_+_34623833 | 0.20 |
ENSDART00000175455
|
si:ch211-269e2.1
|
si:ch211-269e2.1 |
| chr10_+_20299071 | 0.20 |
ENSDART00000111097
|
TCIM (1 of many)
|
chromosome 8 open reading frame 4 |
| chr6_+_33552572 | 0.20 |
ENSDART00000040334
|
pik3r3b
|
phosphoinositide-3-kinase, regulatory subunit 3b (gamma) |
| chr17_-_7990960 | 0.19 |
|
|
|
| chr25_-_22954929 | 0.19 |
ENSDART00000024633
|
dusp8a
|
dual specificity phosphatase 8a |
| chr9_+_41776713 | 0.19 |
|
|
|
| chr25_+_30627892 | 0.19 |
|
|
|
| chr15_-_8333117 | 0.19 |
ENSDART00000061351
|
tnfrsf19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr22_+_38928297 | 0.19 |
ENSDART00000133067
ENSDART00000085701 |
AL953867.2
senp5
|
ENSDARG00000095689 SUMO1/sentrin specific peptidase 5 |
| chr3_-_23340407 | 0.19 |
ENSDART00000113135
ENSDART00000166068 |
msl1a
|
male-specific lethal 1 homolog a (Drosophila) |
| chr5_-_9035835 | 0.19 |
ENSDART00000160079
|
gak
|
cyclin G associated kinase |
| chr21_-_35048238 | 0.19 |
ENSDART00000146454
|
adrb2b
|
adrenoceptor beta 2, surface b |
| chr25_-_26449722 | 0.19 |
ENSDART00000006709
|
dldh
|
dihydrolipoamide dehydrogenase |
| chr15_-_41732304 | 0.19 |
ENSDART00000143447
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr10_+_32739166 | 0.19 |
ENSDART00000063551
|
ppm1e
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr8_+_52429114 | 0.19 |
ENSDART00000021604
|
gins4
|
GINS complex subunit 4 (Sld5 homolog) |
| chr21_+_26954483 | 0.19 |
ENSDART00000065397
|
fkbp2
|
FK506 binding protein 2 |
| chr19_-_35688014 | 0.19 |
|
|
|
| KN150475v1_+_6489 | 0.19 |
|
|
|
| chr9_+_7019681 | 0.19 |
ENSDART00000135576
|
inpp4aa
|
inositol polyphosphate-4-phosphatase type I Aa |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0072578 | neurotransmitter-gated ion channel clustering(GO:0072578) |
| 0.2 | 0.6 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.2 | 0.5 | GO:0097435 | fibril organization(GO:0097435) |
| 0.2 | 0.7 | GO:0046100 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine metabolic process(GO:0046098) guanine biosynthetic process(GO:0046099) hypoxanthine metabolic process(GO:0046100) |
| 0.2 | 0.5 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.1 | 0.4 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.1 | 1.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.6 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.4 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.1 | 0.4 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 1.0 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.6 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.3 | GO:0072526 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.1 | 0.5 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 0.2 | GO:0060544 | regulation of necrotic cell death(GO:0010939) regulation of necroptotic process(GO:0060544) |
| 0.1 | 0.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.3 | GO:2001271 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.2 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 0.2 | GO:0098810 | neurotransmitter uptake(GO:0001504) phasic smooth muscle contraction(GO:0014821) regulation of neurotransmitter uptake(GO:0051580) neurotransmitter reuptake(GO:0098810) |
| 0.1 | 0.8 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 0.2 | GO:0019884 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 0.3 | GO:0040014 | regulation of multicellular organism growth(GO:0040014) |
| 0.1 | 0.5 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.1 | 0.2 | GO:0003085 | negative regulation of systemic arterial blood pressure(GO:0003085) |
| 0.1 | 0.3 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.7 | GO:1902253 | regulation of retinal ganglion cell axon guidance(GO:0090259) regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902253) negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.1 | 0.3 | GO:0072319 | clathrin coat disassembly(GO:0072318) vesicle uncoating(GO:0072319) |
| 0.1 | 1.8 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 0.3 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.1 | 0.3 | GO:0099541 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.1 | 0.6 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.1 | 0.2 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 0.3 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 0.4 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 0.2 | GO:0060031 | protein catabolic process in the vacuole(GO:0007039) mediolateral intercalation(GO:0060031) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) |
| 0.0 | 0.4 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.3 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.1 | GO:0046661 | development of primary male sexual characteristics(GO:0046546) male sex differentiation(GO:0046661) |
| 0.0 | 1.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.4 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.5 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.3 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 0.8 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.0 | 0.4 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.1 | GO:0036073 | perichondral ossification(GO:0036073) |
| 0.0 | 0.3 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.2 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 0.0 | 0.1 | GO:0046888 | negative regulation of hormone secretion(GO:0046888) |
| 0.0 | 0.5 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.1 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.0 | GO:0035552 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.2 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.3 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.8 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.1 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.0 | 0.4 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.4 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.4 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 0.3 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.1 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.2 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.2 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:1902803 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) regulation of synaptic vesicle transport(GO:1902803) |
| 0.0 | 0.1 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.1 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.1 | GO:2001045 | regulation of integrin-mediated signaling pathway(GO:2001044) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.1 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.0 | 0.3 | GO:0015844 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) monoamine transport(GO:0015844) dopamine transport(GO:0015872) catecholamine secretion(GO:0050432) regulation of catecholamine secretion(GO:0050433) catecholamine transport(GO:0051937) regulation of amine transport(GO:0051952) |
| 0.0 | 0.1 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.0 | GO:0071364 | response to epidermal growth factor(GO:0070849) cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.1 | GO:0048532 | anatomical structure arrangement(GO:0048532) |
| 0.0 | 0.1 | GO:0033146 | intracellular estrogen receptor signaling pathway(GO:0030520) regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.0 | 0.3 | GO:0097581 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.0 | 0.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.2 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.0 | GO:0072574 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.0 | 0.2 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.0 | GO:0070476 | rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.0 | 0.2 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.4 | GO:0046890 | regulation of lipid biosynthetic process(GO:0046890) |
| 0.0 | 0.3 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.5 | GO:0051261 | actin filament depolymerization(GO:0030042) protein depolymerization(GO:0051261) |
| 0.0 | 0.2 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:1900182 | positive regulation of protein localization to nucleus(GO:1900182) protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.1 | GO:1901976 | regulation of cell cycle checkpoint(GO:1901976) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 1.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.8 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.7 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.2 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 1.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.3 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.1 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.5 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.6 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.0 | GO:0016234 | inclusion body(GO:0016234) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.2 | 0.7 | GO:0052657 | guanine phosphoribosyltransferase activity(GO:0052657) |
| 0.2 | 0.5 | GO:0003874 | 6-pyruvoyltetrahydropterin synthase activity(GO:0003874) |
| 0.1 | 0.6 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 1.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.8 | GO:0097001 | sphingolipid binding(GO:0046625) ceramide binding(GO:0097001) |
| 0.1 | 0.4 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 0.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.5 | GO:1901611 | phosphatidic acid binding(GO:0070300) phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.1 | 0.4 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.1 | 0.3 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.1 | 0.2 | GO:0001635 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) calcitonin receptor activity(GO:0004948) calcitonin family receptor activity(GO:0097642) |
| 0.1 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.4 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.3 | GO:0016842 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) amidine-lyase activity(GO:0016842) |
| 0.1 | 0.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 1.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.3 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.9 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.0 | 0.5 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.2 | GO:0008515 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.2 | GO:0005536 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.2 | GO:0004935 | adrenergic receptor activity(GO:0004935) |
| 0.0 | 0.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.2 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.3 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.4 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.3 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.0 | 0.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.7 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 1.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.2 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.0 | GO:0035515 | DNA demethylase activity(GO:0035514) oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.0 | GO:0034713 | transforming growth factor beta receptor activity, type II(GO:0005026) type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.6 | GO:0051427 | hormone receptor binding(GO:0051427) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.2 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.0 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.5 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 1.4 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.1 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 1.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.2 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.3 | PID ATM PATHWAY | ATM pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 0.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |