Project
DANIO-CODE
Navigation
Downloads
Results for jund_batf
Z-value: 0.78
Transcription factors associated with jund_batf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
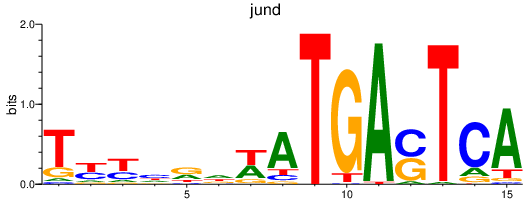
jund
|
ENSDARG00000067850 | JunD proto-oncogene, AP-1 transcription factor subunit |
|
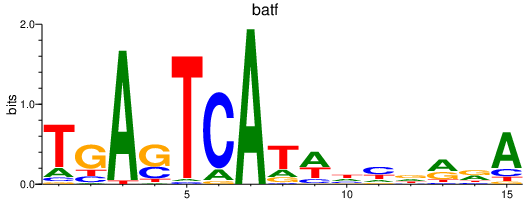
batf
|
ENSDARG00000011818 | basic leucine zipper transcription factor, ATF-like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| jund | dr10_dc_chr2_-_56007484_56007576 | -0.64 | 8.2e-03 | Click! |
| batf | dr10_dc_chr20_+_46668743_46668850 | -0.29 | 2.7e-01 | Click! |
Activity profile of jund_batf motif
Sorted Z-values of jund_batf motif
Network of associatons between targets according to the STRING database.
First level regulatory network of jund_batf
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_24067387 | 1.14 |
ENSDART00000055609
|
atf4b
|
activating transcription factor 4b |
| chr6_+_49096966 | 1.05 |
ENSDART00000141042
ENSDART00000143369 |
slc25a22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr7_+_26730804 | 0.99 |
|
|
|
| chr17_+_989918 | 0.94 |
ENSDART00000169903
|
CABZ01060373.1
|
ENSDARG00000103671 |
| chr22_+_16471319 | 0.92 |
ENSDART00000014330
|
ier5
|
immediate early response 5 |
| chr23_+_6043862 | 0.86 |
|
|
|
| chr7_-_23965038 | 0.85 |
ENSDART00000010124
|
slc7a8a
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8a |
| chr5_-_30324182 | 0.83 |
ENSDART00000153909
|
spns2
|
spinster homolog 2 (Drosophila) |
| chr25_+_18487408 | 0.78 |
ENSDART00000148741
|
met
|
MET proto-oncogene, receptor tyrosine kinase |
| chr3_+_31490043 | 0.77 |
ENSDART00000076636
|
fzd2
|
frizzled class receptor 2 |
| chr15_-_12484651 | 0.71 |
ENSDART00000162258
|
BX901957.1
|
ENSDARG00000098363 |
| chr21_+_22808694 | 0.62 |
ENSDART00000065555
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr21_+_25034997 | 0.61 |
ENSDART00000167523
|
dixdc1b
|
DIX domain containing 1b |
| chr21_+_5781882 | 0.60 |
ENSDART00000091331
|
prodha
|
proline dehydrogenase (oxidase) 1a |
| chr7_-_26165200 | 0.57 |
ENSDART00000123395
|
her8a
|
hairy-related 8a |
| chr4_+_23406078 | 0.57 |
ENSDART00000139543
|
mdm2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr3_-_25886553 | 0.52 |
ENSDART00000030890
|
hmox1a
|
heme oxygenase 1a |
| chr11_-_38940966 | 0.50 |
ENSDART00000105133
|
wnt4a
|
wingless-type MMTV integration site family, member 4a |
| chr25_+_22489716 | 0.49 |
ENSDART00000127831
|
stra6
|
stimulated by retinoic acid 6 |
| chr1_-_22144014 | 0.49 |
ENSDART00000043556
|
ldb2b
|
LIM domain binding 2b |
| chr25_-_18234069 | 0.47 |
ENSDART00000104496
|
dusp6
|
dual specificity phosphatase 6 |
| chr14_-_46648563 | 0.44 |
|
|
|
| chr20_-_22036656 | 0.44 |
ENSDART00000152290
|
daam1b
|
dishevelled associated activator of morphogenesis 1b |
| chr8_+_25880895 | 0.42 |
ENSDART00000124300
|
rhoab
|
ras homolog gene family, member Ab |
| chr23_+_45270759 | 0.41 |
ENSDART00000167056
|
ctc1
|
CTS telomere maintenance complex component 1 |
| chr16_-_13927947 | 0.40 |
ENSDART00000144856
|
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr6_+_56157608 | 0.39 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr13_-_50309241 | 0.38 |
|
|
|
| chr1_-_39368889 | 0.37 |
ENSDART00000136990
|
si:dkey-117m1.4
|
si:dkey-117m1.4 |
| chr21_+_37668334 | 0.36 |
ENSDART00000143621
|
nsd1b
|
nuclear receptor binding SET domain protein 1b |
| chr7_+_44373815 | 0.35 |
ENSDART00000170721
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr14_+_24543732 | 0.35 |
ENSDART00000106039
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr15_-_41288480 | 0.35 |
ENSDART00000155359
|
smco4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr15_+_44260236 | 0.35 |
ENSDART00000128885
|
cwf19l2
|
CWF19-like 2, cell cycle control (S. pombe) |
| chr14_-_17282615 | 0.35 |
ENSDART00000006716
ENSDART00000136242 |
selt2
|
selenoprotein T, 2 |
| chr16_-_13927684 | 0.34 |
ENSDART00000146719
|
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr20_-_22036622 | 0.33 |
ENSDART00000152290
|
daam1b
|
dishevelled associated activator of morphogenesis 1b |
| chr21_+_25729090 | 0.32 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr16_+_25269188 | 0.31 |
ENSDART00000147584
|
hcst
|
hematopoietic cell signal transducer |
| chr8_+_53118919 | 0.31 |
|
|
|
| chr18_+_45121016 | 0.31 |
ENSDART00000172328
|
gyltl1b
|
glycosyltransferase-like 1b |
| chr17_-_23707863 | 0.31 |
ENSDART00000079600
|
minpp1a
|
multiple inositol-polyphosphate phosphatase 1a |
| chr20_-_33802047 | 0.31 |
ENSDART00000166573
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr5_+_69401770 | 0.31 |
|
|
|
| chr20_-_16649213 | 0.31 |
ENSDART00000137601
|
ches1
|
checkpoint suppressor 1 |
| chr23_-_1008307 | 0.28 |
ENSDART00000110588
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr24_+_16998329 | 0.27 |
ENSDART00000177272
|
mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr7_-_54049548 | 0.27 |
ENSDART00000172396
|
fadd
|
Fas (tnfrsf6)-associated via death domain |
| chr20_-_16022955 | 0.26 |
ENSDART00000152357
|
fam20b
|
family with sequence similarity 20, member B (H. sapiens) |
| chr13_+_35213326 | 0.26 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr8_-_985673 | 0.26 |
ENSDART00000170737
|
smyd1b
|
SET and MYND domain containing 1b |
| chr15_-_5575746 | 0.26 |
ENSDART00000130297
|
atg16l2
|
ATG16 autophagy related 16-like 2 (S. cerevisiae) |
| chr16_-_32230355 | 0.26 |
ENSDART00000102027
|
kpna5
|
karyopherin alpha 5 (importin alpha 6) |
| chr23_+_36608246 | 0.26 |
ENSDART00000113179
|
tspan31
|
tetraspanin 31 |
| chr21_-_13571753 | 0.25 |
ENSDART00000065819
|
clic3
|
chloride intracellular channel 3 |
| chr3_-_39346621 | 0.24 |
ENSDART00000135192
ENSDART00000013553 ENSDART00000167289 |
zgc:100868
|
zgc:100868 |
| chr20_-_33802082 | 0.24 |
ENSDART00000166573
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr10_+_36752015 | 0.24 |
ENSDART00000171392
|
rab6a
|
RAB6A, member RAS oncogene family |
| chr8_+_8934762 | 0.23 |
ENSDART00000133137
|
bcap31
|
B-cell receptor-associated protein 31 |
| chr8_-_38168395 | 0.23 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr14_+_30433715 | 0.23 |
ENSDART00000139552
|
atl3
|
atlastin 3 |
| chr13_+_46487890 | 0.23 |
|
|
|
| chr13_-_349685 | 0.23 |
ENSDART00000159803
|
heatr5b
|
HEAT repeat containing 5B |
| chr15_-_44260141 | 0.21 |
ENSDART00000121597
|
CU929391.2
|
ENSDARG00000090304 |
| chr1_-_57842604 | 0.21 |
|
|
|
| chr3_+_15605991 | 0.21 |
ENSDART00000140160
|
BX914211.1
|
ENSDARG00000094116 |
| chr3_-_25886604 | 0.20 |
ENSDART00000030890
|
hmox1a
|
heme oxygenase 1a |
| chr13_-_42986821 | 0.20 |
ENSDART00000164439
|
si:ch211-106f21.1
|
si:ch211-106f21.1 |
| chr10_-_26468280 | 0.20 |
ENSDART00000128894
|
dchs1b
|
dachsous cadherin-related 1b |
| chr8_+_40236694 | 0.20 |
ENSDART00000164414
|
orai1a
|
ORAI calcium release-activated calcium modulator 1a |
| chr5_+_17120453 | 0.19 |
|
|
|
| chr21_+_26684617 | 0.19 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr20_+_23707625 | 0.19 |
|
|
|
| chr2_+_31855153 | 0.19 |
ENSDART00000066789
|
stard3nl
|
STARD3 N-terminal like |
| chr21_-_20674965 | 0.19 |
ENSDART00000065649
|
ENSDARG00000044676
|
ENSDARG00000044676 |
| chr3_+_37565550 | 0.19 |
ENSDART00000151236
|
map3k14a
|
mitogen-activated protein kinase kinase kinase 14a |
| chr19_-_33624795 | 0.19 |
ENSDART00000109868
|
trib1
|
tribbles pseudokinase 1 |
| chr1_+_15827083 | 0.18 |
|
|
|
| chr25_-_10930008 | 0.18 |
ENSDART00000156846
|
mespbb
|
mesoderm posterior bb |
| chr16_-_47491350 | 0.18 |
ENSDART00000149723
|
sept7b
|
septin 7b |
| chr17_+_990022 | 0.18 |
ENSDART00000169903
|
CABZ01060373.1
|
ENSDARG00000103671 |
| chr7_-_39480446 | 0.17 |
ENSDART00000016803
|
grpel1
|
GrpE-like 1, mitochondrial |
| chr13_-_22901327 | 0.17 |
ENSDART00000056523
|
hkdc1
|
hexokinase domain containing 1 |
| chr14_-_46648119 | 0.17 |
|
|
|
| chr14_-_46648801 | 0.17 |
|
|
|
| chr5_+_23585492 | 0.17 |
ENSDART00000135083
|
tp53
|
tumor protein p53 |
| chr15_-_40391882 | 0.16 |
|
|
|
| chr15_-_18179431 | 0.16 |
ENSDART00000047902
|
arcn1b
|
archain 1b |
| chr15_-_5575675 | 0.16 |
ENSDART00000099520
|
atg16l2
|
ATG16 autophagy related 16-like 2 (S. cerevisiae) |
| chr8_+_25015374 | 0.16 |
ENSDART00000140617
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr21_+_37668284 | 0.16 |
ENSDART00000131188
|
nsd1b
|
nuclear receptor binding SET domain protein 1b |
| chr13_+_46899292 | 0.16 |
|
|
|
| chr5_+_53938629 | 0.16 |
ENSDART00000171225
|
npr2
|
natriuretic peptide receptor 2 |
| chr23_+_27830103 | 0.16 |
ENSDART00000137103
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr25_+_25028188 | 0.16 |
ENSDART00000163892
ENSDART00000167542 |
ldha
|
lactate dehydrogenase A4 |
| chr3_+_15208610 | 0.15 |
ENSDART00000141808
|
sh2b1
|
SH2B adaptor protein 1 |
| chr16_-_22394481 | 0.15 |
|
|
|
| chr7_-_35136817 | 0.14 |
ENSDART00000074963
|
lpcat2
|
lysophosphatidylcholine acyltransferase 2 |
| chr7_-_35136743 | 0.14 |
ENSDART00000074963
|
lpcat2
|
lysophosphatidylcholine acyltransferase 2 |
| chr15_+_22458820 | 0.14 |
ENSDART00000109931
|
oafa
|
OAF homolog a (Drosophila) |
| chr25_+_18487313 | 0.14 |
ENSDART00000148741
|
met
|
MET proto-oncogene, receptor tyrosine kinase |
| chr8_+_40236654 | 0.13 |
ENSDART00000164414
|
orai1a
|
ORAI calcium release-activated calcium modulator 1a |
| chr12_-_7604946 | 0.13 |
ENSDART00000126712
|
ccdc6b
|
coiled-coil domain containing 6b |
| chr23_+_1176124 | 0.13 |
|
|
|
| chr16_+_54729975 | 0.12 |
|
|
|
| chr11_-_40414516 | 0.12 |
ENSDART00000165163
|
si:ch211-222l21.1
|
si:ch211-222l21.1 |
| chr4_+_52851409 | 0.12 |
|
|
|
| chr23_+_20936419 | 0.12 |
ENSDART00000129992
|
pax7b
|
paired box 7b |
| chr7_+_6786457 | 0.12 |
ENSDART00000172421
|
rbm14b
|
RNA binding motif protein 14b |
| chr4_+_1680973 | 0.12 |
ENSDART00000149826
ENSDART00000166437 ENSDART00000031135 ENSDART00000172300 ENSDART00000067446 |
slc38a4
|
solute carrier family 38, member 4 |
| chr17_-_15487267 | 0.11 |
|
|
|
| chr18_+_22228682 | 0.10 |
ENSDART00000165464
|
fam65a
|
family with sequence similarity 65, member A |
| chr13_-_10488076 | 0.10 |
ENSDART00000160561
|
si:ch73-54n14.2
|
si:ch73-54n14.2 |
| chr8_+_40236817 | 0.10 |
ENSDART00000164414
|
orai1a
|
ORAI calcium release-activated calcium modulator 1a |
| chr1_+_19376127 | 0.10 |
ENSDART00000088653
|
prss12
|
protease, serine, 12 (neurotrypsin, motopsin) |
| chr19_-_27811300 | 0.10 |
ENSDART00000103940
|
mgat1b
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase b |
| chr12_+_26971596 | 0.10 |
|
|
|
| chr16_+_23367580 | 0.09 |
ENSDART00000015956
|
efna1b
|
ephrin-A1b |
| chr3_+_37433008 | 0.09 |
ENSDART00000055225
|
wnt9b
|
wingless-type MMTV integration site family, member 9B |
| chr21_-_24640528 | 0.09 |
ENSDART00000058370
|
arhgap32b
|
Rho GTPase activating protein 32b |
| chr12_+_26972001 | 0.09 |
|
|
|
| chr19_+_19917767 | 0.08 |
|
|
|
| chr21_-_7702431 | 0.08 |
|
|
|
| chr12_-_10528974 | 0.08 |
ENSDART00000047769
|
myof
|
myoferlin |
| chr13_+_10488748 | 0.08 |
ENSDART00000008603
|
prepl
|
prolyl endopeptidase-like |
| chr21_+_26684728 | 0.07 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr6_-_48318861 | 0.07 |
ENSDART00000103425
|
st7l
|
suppression of tumorigenicity 7 like |
| chr20_-_34767188 | 0.07 |
ENSDART00000033325
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr9_-_23369951 | 0.07 |
|
|
|
| chr23_+_20936374 | 0.07 |
ENSDART00000129992
|
pax7b
|
paired box 7b |
| chr4_-_29350215 | 0.07 |
ENSDART00000169882
|
CR847895.3
|
ENSDARG00000101105 |
| chr12_-_21562929 | 0.07 |
ENSDART00000152999
|
eme1
|
essential meiotic structure-specific endonuclease 1 |
| chr13_+_1741474 | 0.07 |
ENSDART00000161162
|
bmp5
|
bone morphogenetic protein 5 |
| chr20_-_16022816 | 0.06 |
ENSDART00000152828
|
fam20b
|
family with sequence similarity 20, member B (H. sapiens) |
| chr2_-_14758209 | 0.06 |
ENSDART00000162816
|
BX510915.1
|
ENSDARG00000099630 |
| chr6_-_48318769 | 0.06 |
ENSDART00000103425
|
st7l
|
suppression of tumorigenicity 7 like |
| chr3_-_39346658 | 0.06 |
ENSDART00000135192
ENSDART00000013553 ENSDART00000167289 |
zgc:100868
|
zgc:100868 |
| chr13_-_40592360 | 0.06 |
|
|
|
| chr14_+_50394381 | 0.06 |
ENSDART00000158329
|
tnip1
|
TNFAIP3 interacting protein 1 |
| chr23_-_46268313 | 0.06 |
ENSDART00000170417
ENSDART00000168352 |
CABZ01102528.1
|
ENSDARG00000098990 |
| chr23_+_2105366 | 0.06 |
|
|
|
| chr5_+_60871396 | 0.05 |
ENSDART00000131937
|
orai2
|
ORAI calcium release-activated calcium modulator 2 |
| chr12_+_19066653 | 0.05 |
ENSDART00000134726
|
cby1
|
chibby homolog 1 (Drosophila) |
| chr23_+_36607999 | 0.05 |
ENSDART00000113179
|
tspan31
|
tetraspanin 31 |
| chr15_-_39756101 | 0.04 |
|
|
|
| chr17_-_19943800 | 0.04 |
ENSDART00000154251
|
chrm3a
|
cholinergic receptor, muscarinic 3a |
| chr20_+_25441689 | 0.04 |
ENSDART00000063028
|
ctgfa
|
connective tissue growth factor a |
| chr8_-_52759021 | 0.04 |
ENSDART00000168252
|
fgf17
|
fibroblast growth factor 17 |
| chr7_-_37292692 | 0.04 |
ENSDART00000149052
|
nod2
|
nucleotide-binding oligomerization domain containing 2 |
| chr22_-_29387056 | 0.04 |
ENSDART00000121599
|
pdgfba
|
platelet-derived growth factor beta polypeptide a |
| chr2_-_14757989 | 0.04 |
ENSDART00000162816
|
BX510915.1
|
ENSDARG00000099630 |
| chr18_-_5822255 | 0.04 |
ENSDART00000121503
|
cplx3b
|
complexin 3b |
| chr14_-_46649039 | 0.04 |
|
|
|
| chr19_-_27811272 | 0.04 |
ENSDART00000103940
|
mgat1b
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase b |
| chr23_-_1008468 | 0.04 |
ENSDART00000110588
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr9_-_2522639 | 0.04 |
ENSDART00000137706
|
scrn3
|
secernin 3 |
| chr12_+_26971687 | 0.04 |
|
|
|
| chr3_+_15208768 | 0.04 |
ENSDART00000143280
|
sh2b1
|
SH2B adaptor protein 1 |
| chr24_-_11103952 | 0.03 |
|
|
|
| chr23_+_20936600 | 0.03 |
ENSDART00000129992
|
pax7b
|
paired box 7b |
| chr6_-_31380261 | 0.03 |
|
|
|
| chr5_-_3489302 | 0.03 |
|
|
|
| chr14_-_32543646 | 0.02 |
ENSDART00000105726
|
slc25a5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr20_-_31028201 | 0.02 |
ENSDART00000146376
|
acat2
|
acetyl-CoA acetyltransferase 2 |
| chr11_+_7267123 | 0.02 |
ENSDART00000172407
|
si:ch73-238c9.1
|
si:ch73-238c9.1 |
| chr5_-_70915664 | 0.02 |
|
|
|
| chr17_-_19943767 | 0.02 |
ENSDART00000154251
|
chrm3a
|
cholinergic receptor, muscarinic 3a |
| chr15_+_31709362 | 0.02 |
ENSDART00000159634
|
b3glcta
|
beta 3-glucosyltransferase a |
| chr14_-_470112 | 0.02 |
ENSDART00000115093
|
zgc:172215
|
zgc:172215 |
| chr21_-_44570264 | 0.02 |
ENSDART00000159323
|
fundc2
|
fun14 domain containing 2 |
| chr11_+_10552034 | 0.02 |
ENSDART00000157821
|
CT990625.1
|
ENSDARG00000100816 |
| chr13_+_1741432 | 0.02 |
ENSDART00000161162
|
bmp5
|
bone morphogenetic protein 5 |
| chr12_+_30253248 | 0.02 |
ENSDART00000153454
|
ccdc186
|
si:ch211-225b10.4 |
| chr14_-_46648350 | 0.02 |
|
|
|
| chr15_+_22078226 | 0.01 |
ENSDART00000079504
|
ankk1
|
ankyrin repeat and kinase domain containing 1 |
| chr4_+_46917725 | 0.01 |
|
|
|
| chr24_-_24652243 | 0.01 |
ENSDART00000138741
ENSDART00000169581 |
pde7a
|
phosphodiesterase 7A |
| chr16_+_19223683 | 0.01 |
ENSDART00000115263
ENSDART00000114954 |
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr3_-_16056593 | 0.01 |
|
|
|
| chr1_-_24486146 | 0.01 |
ENSDART00000144711
|
tmem154
|
transmembrane protein 154 |
| chr3_+_21539071 | 0.00 |
ENSDART00000156527
|
crhr1
|
corticotropin releasing hormone receptor 1 |
| chr21_-_22914947 | 0.00 |
ENSDART00000083449
|
dub
|
duboraya |
| chr23_+_38314255 | 0.00 |
ENSDART00000129593
|
znf217
|
zinc finger protein 217 |
| chr20_+_53669541 | 0.00 |
ENSDART00000138976
|
bub1ba
|
BUB1 mitotic checkpoint serine/threonine kinase Ba |
| chr7_+_38455040 | 0.00 |
|
|
|
| chr7_+_71262935 | 0.00 |
ENSDART00000100396
|
ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr2_+_31959275 | 0.00 |
ENSDART00000132113
|
otulinb
|
OTU deubiquitinase with linear linkage specificity b |
| chr5_+_26925957 | 0.00 |
|
|
|
| chr5_+_50306662 | 0.00 |
ENSDART00000149892
|
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0043576 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.2 | 0.8 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.2 | 0.6 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.2 | 0.6 | GO:1904019 | epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) |
| 0.2 | 0.5 | GO:1905178 | regulation of cardiac muscle tissue regeneration(GO:1905178) |
| 0.2 | 0.6 | GO:0060544 | regulation of necrotic cell death(GO:0010939) regulation of necroptotic process(GO:0060544) |
| 0.1 | 0.9 | GO:0021707 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.4 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 0.5 | GO:0071939 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.1 | 0.4 | GO:0039703 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.3 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 | 0.8 | GO:0003306 | Wnt signaling pathway involved in heart development(GO:0003306) |
| 0.1 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.2 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.1 | 0.2 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.0 | 0.2 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.0 | 0.3 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.9 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.5 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.2 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.3 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.0 | 0.5 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.4 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.0 | 0.2 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.0 | 0.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0014829 | regulation of smooth muscle contraction(GO:0006940) vascular smooth muscle contraction(GO:0014829) positive regulation of muscle contraction(GO:0045933) |
| 0.0 | 0.4 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.0 | 0.3 | GO:0060729 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.0 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.3 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.4 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 0.1 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.1 | 0.4 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 0.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.3 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 1.0 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.3 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.3 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.3 | GO:0034417 | bisphosphoglycerate 3-phosphatase activity(GO:0034417) |
| 0.1 | 0.5 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.1 | 0.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.5 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 0.9 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.3 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 0.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 0.2 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.5 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.5 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.9 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.1 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.3 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.3 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.6 | GO:0016645 | oxidoreductase activity, acting on the CH-NH group of donors(GO:0016645) |
| 0.0 | 0.3 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.1 | GO:0099528 | G-protein coupled neurotransmitter receptor activity(GO:0099528) |
| 0.0 | 0.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 1.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.0 | GO:0050700 | CARD domain binding(GO:0050700) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.9 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.3 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 0.9 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 0.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 0.6 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.0 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |