Project
DANIO-CODE
Navigation
Downloads
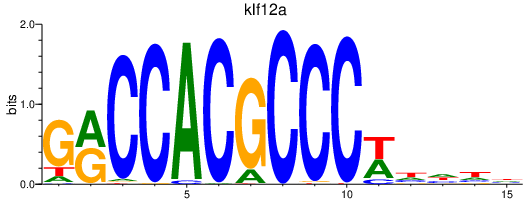
Results for klf12a
Z-value: 0.78
Transcription factors associated with klf12a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
klf12a
|
ENSDARG00000015312 | Kruppel-like factor 12a |
Activity profile of klf12a motif
Sorted Z-values of klf12a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of klf12a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_53568814 | 1.73 |
ENSDART00000154189
|
rbm24b
|
RNA binding motif protein 24b |
| chr14_+_6742208 | 1.18 |
ENSDART00000109138
|
hbegfa
|
heparin-binding EGF-like growth factor a |
| chr23_-_31719203 | 1.14 |
ENSDART00000148122
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr8_-_410733 | 1.07 |
ENSDART00000151255
|
trim36
|
tripartite motif containing 36 |
| chr8_+_23192085 | 1.05 |
ENSDART00000032996
ENSDART00000137536 |
ppdpfa
|
pancreatic progenitor cell differentiation and proliferation factor a |
| chr13_-_49886891 | 1.04 |
ENSDART00000074230
|
pkz
|
protein kinase containing Z-DNA binding domains |
| chr13_-_36495730 | 0.92 |
ENSDART00000165629
|
cdkn3
|
cyclin-dependent kinase inhibitor 3 |
| chr17_-_18878230 | 0.88 |
ENSDART00000028044
|
galcb
|
galactosylceramidase b |
| chr22_+_818795 | 0.82 |
ENSDART00000003325
|
dennd2db
|
DENN/MADD domain containing 2Db |
| chr25_+_7318688 | 0.82 |
ENSDART00000166496
|
cat
|
catalase |
| chr3_-_52644257 | 0.75 |
ENSDART00000128223
|
CHCHD5
|
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr5_+_2472901 | 0.75 |
ENSDART00000172699
|
gng10
|
guanine nucleotide binding protein (G protein), gamma 10 |
| chr3_+_54551887 | 0.75 |
ENSDART00000169663
|
CT573860.1
|
ENSDARG00000098327 |
| chr8_-_32376710 | 0.70 |
ENSDART00000098850
|
lipg
|
lipase, endothelial |
| chr23_-_17503357 | 0.68 |
ENSDART00000043076
|
ppdpfb
|
pancreatic progenitor cell differentiation and proliferation factor b |
| chr25_+_5995507 | 0.66 |
ENSDART00000067514
|
rbpms2a
|
RNA binding protein with multiple splicing 2a |
| chr2_+_25904910 | 0.66 |
ENSDART00000133623
|
pld1a
|
phospholipase D1a |
| chr23_+_30971437 | 0.65 |
ENSDART00000146859
|
cables2a
|
Cdk5 and Abl enzyme substrate 2a |
| chr16_-_17289585 | 0.63 |
ENSDART00000135146
|
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr20_-_15205171 | 0.63 |
ENSDART00000034011
|
fmo5
|
flavin containing monooxygenase 5 |
| chr17_+_25314291 | 0.63 |
ENSDART00000082319
|
tmem54a
|
transmembrane protein 54a |
| chr10_-_34058331 | 0.62 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr10_+_19597171 | 0.62 |
ENSDART00000063810
ENSDART00000063806 |
atp6v1b2
|
ATPase, H+ transporting, lysosomal, V1 subunit B2 |
| chr22_+_24596299 | 0.61 |
ENSDART00000158303
ENSDART00000160924 |
mcoln2
|
mucolipin 2 |
| chr19_+_47698663 | 0.60 |
ENSDART00000161845
ENSDART00000078382 ENSDART00000158262 |
tpmt.1
|
thiopurine S-methyltransferase, tandem duplicate 1 |
| chr19_+_46523185 | 0.59 |
ENSDART00000159331
|
rbm24a
|
RNA binding motif protein 24a |
| chr16_-_26258657 | 0.57 |
ENSDART00000157787
|
lipeb
|
lipase, hormone-sensitive b |
| chr14_+_11803542 | 0.57 |
ENSDART00000121913
|
kctd12b
|
potassium channel tetramerisation domain containing 12b |
| chr6_+_41506220 | 0.56 |
ENSDART00000136538
|
cish
|
cytokine inducible SH2-containing protein |
| chr10_+_40031949 | 0.56 |
ENSDART00000009857
|
ENSDARG00000003145
|
ENSDARG00000003145 |
| chr9_+_30610756 | 0.55 |
ENSDART00000145025
|
zgc:113314
|
zgc:113314 |
| chr13_-_33562119 | 0.54 |
ENSDART00000143945
ENSDART00000100504 |
zgc:163030
|
zgc:163030 |
| chr10_+_7678087 | 0.53 |
ENSDART00000160673
|
hint1
|
histidine triad nucleotide binding protein 1 |
| chr21_-_13759788 | 0.52 |
ENSDART00000102197
|
fam129ba
|
family with sequence similarity 129, member Ba |
| chr6_+_18024734 | 0.52 |
ENSDART00000067560
|
acox1
|
acyl-CoA oxidase 1, palmitoyl |
| chr25_+_25642475 | 0.51 |
ENSDART00000108476
|
lrrc61
|
leucine rich repeat containing 61 |
| chr5_+_36250811 | 0.50 |
ENSDART00000051186
|
nccrp1
|
non-specific cytotoxic cell receptor protein 1 |
| chr10_-_42302932 | 0.50 |
ENSDART00000076693
ENSDART00000073631 |
stambpa
|
STAM binding protein a |
| chr5_-_11530880 | 0.49 |
ENSDART00000159896
|
gatsl3
|
GATS protein-like 3 |
| chr10_-_20681689 | 0.49 |
|
|
|
| chr7_-_3332675 | 0.48 |
|
|
|
| chr7_-_58427369 | 0.48 |
ENSDART00000149347
|
chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr11_+_44909949 | 0.48 |
ENSDART00000173116
ENSDART00000167011 ENSDART00000167226 |
pycr1b
|
pyrroline-5-carboxylate reductase 1b |
| chr11_-_40383013 | 0.48 |
ENSDART00000112140
|
fam213b
|
family with sequence similarity 213, member B |
| chr21_-_32027717 | 0.47 |
ENSDART00000131651
|
ENSDARG00000073961
|
ENSDARG00000073961 |
| chr14_-_136402 | 0.46 |
|
|
|
| chr11_-_11317608 | 0.46 |
ENSDART00000113311
|
col9a1a
|
collagen, type IX, alpha 1a |
| chr17_-_7635061 | 0.46 |
ENSDART00000064655
|
zbtb2a
|
zinc finger and BTB domain containing 2a |
| chr11_-_26595578 | 0.46 |
ENSDART00000153519
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr21_+_21242470 | 0.45 |
ENSDART00000148346
|
itpkca
|
inositol-trisphosphate 3-kinase Ca |
| chr18_+_38210277 | 0.45 |
ENSDART00000052703
|
nucb2b
|
nucleobindin 2b |
| chr11_-_34314863 | 0.45 |
ENSDART00000133302
|
pfkfb4a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4a |
| chr22_-_3747016 | 0.44 |
ENSDART00000083308
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr17_+_25314229 | 0.44 |
ENSDART00000082319
|
tmem54a
|
transmembrane protein 54a |
| chr9_-_28588288 | 0.44 |
ENSDART00000104317
ENSDART00000064343 |
klf7b
|
Kruppel-like factor 7b |
| chr6_+_41506350 | 0.44 |
ENSDART00000136538
|
cish
|
cytokine inducible SH2-containing protein |
| chr7_-_58856734 | 0.44 |
ENSDART00000159285
|
haus6
|
HAUS augmin-like complex, subunit 6 |
| chr18_-_6336677 | 0.44 |
ENSDART00000169844
|
iqsec3b
|
IQ motif and Sec7 domain 3b |
| chr7_+_38690837 | 0.44 |
ENSDART00000173565
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr15_-_19192862 | 0.43 |
ENSDART00000152428
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr25_+_25642290 | 0.43 |
ENSDART00000108476
|
lrrc61
|
leucine rich repeat containing 61 |
| chr7_-_58856862 | 0.43 |
ENSDART00000164104
|
haus6
|
HAUS augmin-like complex, subunit 6 |
| chr11_+_29543388 | 0.43 |
ENSDART00000067822
|
dynlt3
|
dynein, light chain, Tctex-type 3 |
| chr7_+_38490753 | 0.43 |
ENSDART00000036461
|
arhgap1
|
Rho GTPase activating protein 1 |
| chr11_+_25063081 | 0.42 |
|
|
|
| chr20_-_43889302 | 0.42 |
ENSDART00000004601
|
laptm4a
|
lysosomal protein transmembrane 4 alpha |
| chr7_-_58856806 | 0.42 |
ENSDART00000159285
|
haus6
|
HAUS augmin-like complex, subunit 6 |
| chr2_+_30932612 | 0.41 |
ENSDART00000132450
ENSDART00000137012 |
myom1a
|
myomesin 1a (skelemin) |
| chr1_+_45230899 | 0.41 |
ENSDART00000033669
|
lipt1
|
lipoyltransferase 1 |
| chr16_+_19831573 | 0.41 |
ENSDART00000135359
|
macc1
|
metastasis associated in colon cancer 1 |
| chr8_-_4562273 | 0.40 |
ENSDART00000025535
|
sept5a
|
septin 5a |
| chr5_-_9320252 | 0.40 |
ENSDART00000169698
|
zgc:158343
|
zgc:158343 |
| chr7_-_73592082 | 0.40 |
ENSDART00000160951
|
hist1h4l
|
histone 1, H4, like |
| chr10_+_3299541 | 0.40 |
ENSDART00000031121
|
ENSDARG00000021564
|
ENSDARG00000021564 |
| KN149795v1_-_10712 | 0.39 |
|
|
|
| chr13_-_36496048 | 0.39 |
ENSDART00000057155
|
cdkn3
|
cyclin-dependent kinase inhibitor 3 |
| chr25_+_18910528 | 0.39 |
ENSDART00000104420
|
samm50
|
SAMM50 sorting and assembly machinery component |
| chr19_+_30266913 | 0.38 |
ENSDART00000109729
|
si:ch73-130a3.4
|
si:ch73-130a3.4 |
| chr6_+_41506504 | 0.37 |
ENSDART00000084861
|
cish
|
cytokine inducible SH2-containing protein |
| chr1_+_45015350 | 0.37 |
ENSDART00000143363
|
si:ch211-214c7.4
|
si:ch211-214c7.4 |
| chr17_+_51675406 | 0.37 |
ENSDART00000157171
|
si:ch211-168d23.3
|
si:ch211-168d23.3 |
| chr22_-_20899539 | 0.37 |
ENSDART00000100642
|
ell
|
elongation factor RNA polymerase II |
| chr22_-_38271070 | 0.37 |
ENSDART00000165430
ENSDART00000059690 |
hps3
|
Hermansky-Pudlak syndrome 3 |
| chr8_-_17132057 | 0.37 |
ENSDART00000143920
|
mrps36
|
mitochondrial ribosomal protein S36 |
| chr10_+_33439121 | 0.37 |
ENSDART00000063662
|
mdh2
|
malate dehydrogenase 2, NAD (mitochondrial) |
| chr9_-_7676791 | 0.36 |
ENSDART00000136438
|
dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr10_-_17753753 | 0.36 |
|
|
|
| chr17_-_4160288 | 0.36 |
ENSDART00000153824
|
napba
|
N-ethylmaleimide-sensitive factor attachment protein, beta a |
| chr18_+_44656426 | 0.36 |
ENSDART00000059063
|
ehd2b
|
EH-domain containing 2b |
| chr17_+_44327269 | 0.35 |
ENSDART00000045882
|
ap5m1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr6_+_59762717 | 0.34 |
ENSDART00000050457
|
zgc:65895
|
zgc:65895 |
| chr20_-_194135 | 0.34 |
ENSDART00000064546
|
si:ch211-241j12.3
|
si:ch211-241j12.3 |
| chr11_-_37816180 | 0.34 |
ENSDART00000086516
|
klhdc8a
|
kelch domain containing 8A |
| chr21_+_6091802 | 0.34 |
ENSDART00000147716
|
si:dkey-93m18.3
|
si:dkey-93m18.3 |
| chr4_+_9010972 | 0.34 |
ENSDART00000058007
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr1_-_689420 | 0.34 |
|
|
|
| chr6_+_41506446 | 0.33 |
ENSDART00000140108
|
cish
|
cytokine inducible SH2-containing protein |
| chr7_+_28773716 | 0.33 |
ENSDART00000086871
|
gfod2
|
glucose-fructose oxidoreductase domain containing 2 |
| chr23_-_17543731 | 0.33 |
ENSDART00000002398
|
trim101
|
tripartite motif containing 101 |
| chr5_-_3673698 | 0.33 |
|
|
|
| chr23_+_30971404 | 0.33 |
ENSDART00000146859
|
cables2a
|
Cdk5 and Abl enzyme substrate 2a |
| chr21_+_1622040 | 0.33 |
ENSDART00000148540
|
fech
|
ferrochelatase |
| chr16_+_5542562 | 0.33 |
|
|
|
| chr9_-_7676688 | 0.33 |
ENSDART00000102706
|
dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr11_-_36017410 | 0.33 |
ENSDART00000122531
|
nfya
|
nuclear transcription factor Y, alpha |
| chr9_-_296560 | 0.33 |
ENSDART00000160220
|
kif5aa
|
kinesin family member 5A, a |
| chr25_+_4414750 | 0.33 |
ENSDART00000129978
|
pnpla2
|
patatin-like phospholipase domain containing 2 |
| chr10_-_44170848 | 0.32 |
ENSDART00000135240
|
acads
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| KN150123v1_-_37214 | 0.32 |
|
|
|
| chr5_+_36456679 | 0.32 |
ENSDART00000036760
|
tppp2
|
tubulin polymerization-promoting protein family member 2 |
| chr8_+_53173077 | 0.32 |
ENSDART00000131514
|
nadka
|
NAD kinase a |
| chr18_-_33832 | 0.31 |
ENSDART00000158957
ENSDART00000129125 |
pde8a
|
phosphodiesterase 8A |
| chr3_+_14238188 | 0.31 |
ENSDART00000165452
ENSDART00000171726 |
tmem56b
|
transmembrane protein 56b |
| chr25_-_36713728 | 0.31 |
ENSDART00000103222
|
nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr4_+_9010890 | 0.31 |
ENSDART00000058007
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr7_+_23636549 | 0.31 |
ENSDART00000045479
|
syt4
|
synaptotagmin IV |
| chr22_-_12601276 | 0.31 |
ENSDART00000143261
|
cep70
|
centrosomal protein 70 |
| chr25_-_5921772 | 0.31 |
ENSDART00000075184
|
snx1a
|
sorting nexin 1a |
| chr4_-_4603294 | 0.31 |
ENSDART00000130601
|
CABZ01020840.1
|
ENSDARG00000090401 |
| chr24_-_10935904 | 0.30 |
ENSDART00000003195
|
chmp4c
|
charged multivesicular body protein 4C |
| chr11_-_42263721 | 0.30 |
ENSDART00000130573
|
atp6ap1la
|
ATPase, H+ transporting, lysosomal accessory protein 1-like a |
| chr25_+_7318825 | 0.30 |
ENSDART00000159748
|
cat
|
catalase |
| chr18_+_24490621 | 0.30 |
ENSDART00000143108
|
CR847971.1
|
ENSDARG00000091941 |
| chr18_+_27095471 | 0.29 |
ENSDART00000125326
ENSDART00000098334 |
ppp1r15b
|
protein phosphatase 1, regulatory subunit 15B |
| chr9_-_1983772 | 0.29 |
ENSDART00000082339
|
hoxd12a
|
homeobox D12a |
| chr18_+_6417959 | 0.29 |
ENSDART00000110892
|
b4galnt3b
|
beta-1,4-N-acetyl-galactosaminyl transferase 3b |
| chr22_-_11817641 | 0.29 |
ENSDART00000000192
|
ptpn4b
|
protein tyrosine phosphatase, non-receptor type 4b |
| chr23_+_14834670 | 0.29 |
ENSDART00000143675
|
si:rp71-79p20.2
|
si:rp71-79p20.2 |
| KN150165v1_+_30233 | 0.29 |
|
|
|
| chr23_-_17503329 | 0.29 |
ENSDART00000132024
|
ppdpfb
|
pancreatic progenitor cell differentiation and proliferation factor b |
| chr2_+_23567197 | 0.29 |
ENSDART00000167068
|
kifap3b
|
kinesin-associated protein 3b |
| chr20_-_15205248 | 0.29 |
ENSDART00000034011
|
fmo5
|
flavin containing monooxygenase 5 |
| chr21_-_22085596 | 0.29 |
ENSDART00000101726
|
slc35f2
|
solute carrier family 35, member F2 |
| chr14_+_4044357 | 0.28 |
ENSDART00000160431
|
dhrs13l1
|
dehydrogenase/reductase (SDR family) member 13 like 1 |
| chr7_+_28881845 | 0.28 |
ENSDART00000138065
|
tmppe
|
transmembrane protein with metallophosphoesterase domain |
| chr19_+_47707575 | 0.28 |
ENSDART00000163591
ENSDART00000159860 ENSDART00000138909 ENSDART00000158588 ENSDART00000170847 |
tpmt.2
|
thiopurine S-methyltransferase, tandem duplicate 2 |
| chr20_+_1175551 | 0.28 |
ENSDART00000041192
|
lyrm2
|
LYR motif containing 2 |
| chr16_+_5377008 | 0.28 |
ENSDART00000133972
|
plecb
|
plectin b |
| chr6_+_54070547 | 0.27 |
ENSDART00000110845
|
fhit
|
fragile histidine triad gene |
| chr2_-_56531858 | 0.27 |
ENSDART00000113589
|
gpx4b
|
glutathione peroxidase 4b |
| chr22_-_11817587 | 0.27 |
ENSDART00000000192
|
ptpn4b
|
protein tyrosine phosphatase, non-receptor type 4b |
| chr9_+_28420902 | 0.27 |
|
|
|
| chr10_-_41430693 | 0.27 |
ENSDART00000052971
ENSDART00000128156 |
rab11fip1b
|
RAB11 family interacting protein 1 (class I) b |
| chr7_-_10318801 | 0.27 |
ENSDART00000172761
ENSDART00000170476 |
mthfs
|
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chr4_-_4603205 | 0.27 |
ENSDART00000130601
|
CABZ01020840.1
|
ENSDARG00000090401 |
| chr9_+_28420988 | 0.27 |
|
|
|
| chr20_-_54591757 | 0.27 |
ENSDART00000136779
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr23_-_10810264 | 0.26 |
ENSDART00000013768
|
eif4e3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr10_-_625501 | 0.26 |
ENSDART00000041236
ENSDART00000144882 |
rfx3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr10_+_37457234 | 0.26 |
ENSDART00000136554
|
akap1a
|
A kinase (PRKA) anchor protein 1a |
| chr13_+_36638875 | 0.26 |
ENSDART00000111832
|
atl1
|
atlastin GTPase 1 |
| chr25_-_25641897 | 0.26 |
ENSDART00000166308
|
cib2
|
calcium and integrin binding family member 2 |
| chr11_-_6870810 | 0.26 |
ENSDART00000007204
|
ddx49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr4_-_18660286 | 0.26 |
ENSDART00000178638
|
pparaa
|
peroxisome proliferator-activated receptor alpha a |
| chr15_-_43283562 | 0.26 |
ENSDART00000110352
|
ap1s3a
|
adaptor-related protein complex 1, sigma 3 subunit, a |
| chr6_+_29702868 | 0.25 |
ENSDART00000114172
|
pde6d
|
phosphodiesterase 6D, cGMP-specific, rod, delta |
| chr2_+_19588034 | 0.25 |
ENSDART00000163875
|
cc2d1b
|
coiled-coil and C2 domain containing 1B |
| chr9_-_296499 | 0.25 |
ENSDART00000165228
|
kif5aa
|
kinesin family member 5A, a |
| chr25_+_22222388 | 0.25 |
ENSDART00000154376
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr14_+_26141096 | 0.25 |
ENSDART00000054183
|
gpr137
|
G protein-coupled receptor 137 |
| chr14_+_32923977 | 0.25 |
ENSDART00000075187
|
pdzd11
|
PDZ domain containing 11 |
| chr12_-_33481348 | 0.25 |
ENSDART00000009794
ENSDART00000136617 |
tdrkh
|
tudor and KH domain containing |
| chr21_+_13063614 | 0.25 |
|
|
|
| chr2_+_26002426 | 0.25 |
ENSDART00000051234
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr12_+_42281154 | 0.25 |
ENSDART00000167324
|
ebf3a
|
early B-cell factor 3a |
| chr21_-_43332911 | 0.25 |
ENSDART00000114955
|
sowahaa
|
sosondowah ankyrin repeat domain family member Aa |
| chr17_+_16421892 | 0.24 |
ENSDART00000136078
|
efcab11
|
EF-hand calcium binding domain 11 |
| chr10_+_37556367 | 0.24 |
ENSDART00000135642
|
msi2a
|
musashi RNA-binding protein 2a |
| chr15_-_30935719 | 0.24 |
ENSDART00000050649
|
msi2b
|
musashi RNA-binding protein 2b |
| chr15_-_19192933 | 0.24 |
ENSDART00000152428
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr15_-_9616877 | 0.24 |
ENSDART00000168668
|
gab2
|
GRB2-associated binding protein 2 |
| chr16_+_14135089 | 0.24 |
|
|
|
| chr19_+_8225252 | 0.24 |
ENSDART00000147218
|
efna3a
|
ephrin-A3a |
| chr7_-_6295302 | 0.24 |
ENSDART00000173199
|
HIST2H2AB (1 of many)
|
si:ch1073-153i20.5 |
| chr1_-_6403371 | 0.23 |
|
|
|
| chr17_-_36988937 | 0.23 |
ENSDART00000145236
|
dpysl5a
|
dihydropyrimidinase-like 5a |
| chr13_+_16148867 | 0.23 |
ENSDART00000142408
|
anxa11a
|
annexin A11a |
| chr25_-_13057808 | 0.23 |
ENSDART00000172571
|
smpd3
|
sphingomyelin phosphodiesterase 3, neutral |
| chr13_-_31267133 | 0.23 |
ENSDART00000008287
|
pgam1a
|
phosphoglycerate mutase 1a |
| chr1_-_45972732 | 0.23 |
ENSDART00000134450
|
ebpl
|
emopamil binding protein-like |
| chr1_+_45015381 | 0.23 |
ENSDART00000143363
|
si:ch211-214c7.4
|
si:ch211-214c7.4 |
| chr25_+_4508802 | 0.23 |
ENSDART00000021120
|
pgghg
|
protein-glucosylgalactosylhydroxylysine glucosidase |
| chr4_-_4698425 | 0.22 |
ENSDART00000093005
|
ENSDARG00000063578
|
ENSDARG00000063578 |
| chr8_-_410190 | 0.22 |
ENSDART00000151155
|
trim36
|
tripartite motif containing 36 |
| chr6_-_40660116 | 0.22 |
ENSDART00000154359
|
ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr23_-_10810190 | 0.22 |
ENSDART00000140745
|
eif4e3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr10_-_17753457 | 0.22 |
ENSDART00000147553
|
stard7
|
StAR-related lipid transfer (START) domain containing 7 |
| chr15_+_28477741 | 0.22 |
ENSDART00000057257
|
pitpnaa
|
phosphatidylinositol transfer protein, alpha a |
| chr5_-_11531015 | 0.22 |
ENSDART00000159896
|
gatsl3
|
GATS protein-like 3 |
| chr7_+_10320888 | 0.21 |
ENSDART00000173125
|
zfand6
|
zinc finger, AN1-type domain 6 |
| chr3_+_17801981 | 0.21 |
ENSDART00000164663
|
aclya
|
ATP citrate lyase a |
| chr2_+_11245538 | 0.21 |
ENSDART00000138737
ENSDART00000081058 |
acot11a
|
acyl-CoA thioesterase 11a |
| chr13_-_42547873 | 0.21 |
ENSDART00000169083
|
lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr25_+_3381667 | 0.21 |
ENSDART00000157777
|
zgc:153293
|
zgc:153293 |
| chr6_+_22049537 | 0.21 |
ENSDART00000158263
|
si:dkey-10p5.10
|
si:dkey-10p5.10 |
| chr16_-_52847642 | 0.21 |
ENSDART00000147236
|
azin1a
|
antizyme inhibitor 1a |
| chr2_-_1677190 | 0.21 |
ENSDART00000024135
|
tubb2
|
tubulin, beta 2A class IIa |
| chr5_+_37662896 | 0.21 |
ENSDART00000051231
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr3_-_36117170 | 0.21 |
ENSDART00000158701
ENSDART00000141106 |
gps1
|
G protein pathway suppressor 1 |
| chr13_-_42548129 | 0.21 |
ENSDART00000169083
|
lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.3 | 1.0 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.3 | 0.9 | GO:0006683 | galactosylceramide catabolic process(GO:0006683) galactolipid catabolic process(GO:0019376) |
| 0.3 | 1.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.3 | 0.3 | GO:0015961 | diadenosine polyphosphate metabolic process(GO:0015959) diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.2 | 1.7 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.2 | 0.6 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.2 | 0.5 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.2 | 0.7 | GO:0051150 | regulation of smooth muscle cell differentiation(GO:0051150) negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 1.2 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.1 | 0.7 | GO:0032534 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.3 | GO:0071364 | response to epidermal growth factor(GO:0070849) cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.1 | 0.7 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.1 | 0.5 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.4 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 0.3 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 1.1 | GO:0042542 | response to hydrogen peroxide(GO:0042542) |
| 0.1 | 0.3 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.1 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.4 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 0.3 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) protein demalonylation(GO:0036046) peptidyl-lysine demalonylation(GO:0036047) protein desuccinylation(GO:0036048) peptidyl-lysine desuccinylation(GO:0036049) protein deglutarylation(GO:0061698) |
| 0.1 | 0.2 | GO:0015846 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 0.4 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 0.2 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 0.3 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.4 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.2 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 0.5 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 0.4 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.2 | GO:0046661 | development of primary male sexual characteristics(GO:0046546) male sex differentiation(GO:0046661) |
| 0.0 | 0.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.5 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.0 | 0.9 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.8 | GO:0009154 | purine ribonucleotide catabolic process(GO:0009154) |
| 0.0 | 0.2 | GO:0010922 | positive regulation of phosphatase activity(GO:0010922) positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.3 | GO:0010888 | negative regulation of cholesterol storage(GO:0010887) negative regulation of lipid storage(GO:0010888) positive regulation of fatty acid metabolic process(GO:0045923) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.3 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 1.0 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.2 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 0.3 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 1.3 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.1 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.4 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.5 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.5 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.5 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.3 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.2 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.1 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.0 | 0.2 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 1.2 | GO:0031017 | exocrine pancreas development(GO:0031017) exocrine system development(GO:0035272) |
| 0.0 | 0.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.1 | GO:0060043 | regulation of cardiac muscle tissue growth(GO:0055021) regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.0 | 0.2 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.0 | 0.1 | GO:0002551 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.6 | GO:0018198 | peptidyl-cysteine modification(GO:0018198) |
| 0.0 | 0.3 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.5 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.2 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 1.3 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.0 | 0.0 | GO:0052805 | imidazole-containing compound metabolic process(GO:0052803) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.2 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.2 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.3 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.4 | GO:0043648 | dicarboxylic acid metabolic process(GO:0043648) |
| 0.0 | 0.2 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.8 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.9 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.1 | GO:1902743 | regulation of lamellipodium assembly(GO:0010591) regulation of lamellipodium organization(GO:1902743) |
| 0.0 | 0.0 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 0.3 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.7 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.5 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.4 | GO:0015914 | phospholipid transport(GO:0015914) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 1.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.4 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.4 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.1 | 0.9 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 0.7 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.2 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.5 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.4 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 1.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.1 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 1.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 1.5 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.3 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.5 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.5 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.4 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0004336 | galactosylceramidase activity(GO:0004336) |
| 0.2 | 1.0 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.2 | 0.9 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 0.7 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.2 | 0.5 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 0.6 | GO:1903231 | mRNA binding involved in posttranscriptional gene silencing(GO:1903231) |
| 0.1 | 0.6 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.6 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.5 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.7 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.3 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.1 | 0.9 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 0.3 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.1 | 0.4 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.3 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 0.3 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.1 | 1.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.3 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 0.3 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 0.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.7 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.3 | GO:0061697 | protein-malonyllysine demalonylase activity(GO:0036054) protein-succinyllysine desuccinylase activity(GO:0036055) protein-glutaryllysine deglutarylase activity(GO:0061697) |
| 0.1 | 0.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 0.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 1.1 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 0.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.4 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 1.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.2 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.5 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 1.1 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.3 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.3 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.7 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.5 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0008988 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) |
| 0.0 | 0.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 2.3 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 1.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.2 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 2.3 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.5 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 1.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 1.7 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 0.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 0.6 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 1.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.4 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |