Project
DANIO-CODE
Navigation
Downloads
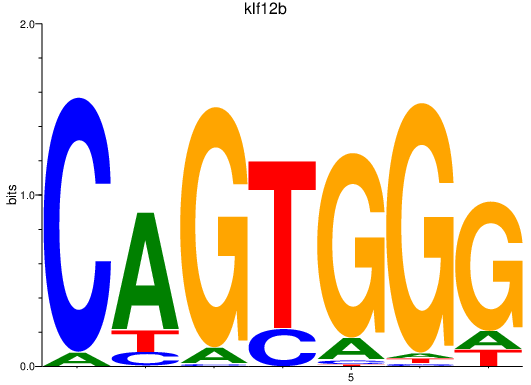
Results for klf12b
Z-value: 3.27
Transcription factors associated with klf12b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
klf12b
|
ENSDARG00000032197 | Kruppel-like factor 12b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| klf12b | dr10_dc_chr9_+_30909382_30909427 | 0.94 | 7.7e-08 | Click! |
Activity profile of klf12b motif
Sorted Z-values of klf12b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of klf12b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_43501747 | 10.45 |
ENSDART00000002279
|
cdh11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr13_-_39821399 | 9.37 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr20_+_17840364 | 9.19 |
ENSDART00000024627
|
cdh2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr16_+_24057260 | 8.25 |
ENSDART00000132742
ENSDART00000145330 |
apoc1
|
apolipoprotein C-I |
| chr25_-_30845998 | 8.13 |
ENSDART00000027661
|
myod1
|
myogenic differentiation 1 |
| chr13_+_13550656 | 7.72 |
ENSDART00000057825
|
cfd
|
complement factor D (adipsin) |
| chr22_-_4264838 | 7.51 |
ENSDART00000125302
|
fbn2b
|
fibrillin 2b |
| chr6_+_57744081 | 6.46 |
ENSDART00000169175
|
SNTA1
|
syntrophin alpha 1 |
| chr22_-_26333957 | 6.35 |
ENSDART00000130493
|
capn2b
|
calpain 2, (m/II) large subunit b |
| chr7_+_44373815 | 6.26 |
ENSDART00000170721
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr2_-_20941256 | 6.17 |
ENSDART00000114199
|
si:ch211-267e7.3
|
si:ch211-267e7.3 |
| chr13_-_11404389 | 6.14 |
ENSDART00000018155
|
adss
|
adenylosuccinate synthase |
| chr21_-_37509454 | 6.02 |
ENSDART00000175126
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr22_+_12406830 | 5.77 |
ENSDART00000108609
|
rnd3a
|
Rho family GTPase 3a |
| chr20_-_19646761 | 5.69 |
|
|
|
| KN150256v1_-_9467 | 5.44 |
|
|
|
| chr5_-_66792947 | 5.42 |
ENSDART00000147009
|
si:dkey-251i10.2
|
si:dkey-251i10.2 |
| chr20_+_23392782 | 5.42 |
|
|
|
| chr16_+_24032160 | 5.37 |
ENSDART00000103190
|
apoa4b.2
|
apolipoprotein A-IV b, tandem duplicate 2 |
| chr6_+_19795100 | 5.35 |
|
|
|
| chr2_-_29501573 | 5.31 |
ENSDART00000138073
|
cahz
|
carbonic anhydrase |
| chr5_+_51392394 | 5.19 |
ENSDART00000143276
|
setbp1
|
SET binding protein 1 |
| chr17_-_49878964 | 5.19 |
ENSDART00000154728
|
col12a1a
|
collagen, type XII, alpha 1a |
| chr7_-_43501636 | 5.17 |
ENSDART00000002279
|
cdh11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr19_-_4079427 | 5.10 |
ENSDART00000170325
|
map7d1b
|
MAP7 domain containing 1b |
| chr22_-_2870591 | 5.08 |
ENSDART00000063533
|
aqp12
|
aquaporin 12 |
| chr1_+_16683931 | 4.99 |
ENSDART00000103262
ENSDART00000145068 ENSDART00000169619 ENSDART00000010526 |
fat1a
|
FAT atypical cadherin 1a |
| chr3_+_15355472 | 4.87 |
ENSDART00000141714
|
nupr1
|
nuclear protein 1 |
| chr1_+_26418707 | 4.86 |
ENSDART00000161169
|
bnc2
|
basonuclin 2 |
| chr22_+_5697877 | 4.86 |
ENSDART00000063484
|
si:dkey-222f2.1
|
si:dkey-222f2.1 |
| chr24_-_31772736 | 4.86 |
|
|
|
| chr14_-_46219646 | 4.84 |
ENSDART00000110191
|
shisa3
|
shisa family member 3 |
| chr23_-_9924987 | 4.77 |
ENSDART00000005015
|
prkcbp1l
|
protein kinase C binding protein 1, like |
| chr25_-_2485276 | 4.74 |
ENSDART00000154889
ENSDART00000155027 |
BX957346.1
|
ENSDARG00000096850 |
| chr1_+_44137973 | 4.68 |
|
|
|
| chr11_-_471166 | 4.67 |
ENSDART00000154888
|
cnbpb
|
CCHC-type zinc finger, nucleic acid binding protein b |
| chr7_-_69639922 | 4.66 |
|
|
|
| chr14_-_20760787 | 4.60 |
|
|
|
| chr14_+_20809272 | 4.58 |
ENSDART00000139865
|
aldob
|
aldolase b, fructose-bisphosphate |
| chr11_+_23695123 | 4.56 |
ENSDART00000000486
|
cntn2
|
contactin 2 |
| chr2_+_19546837 | 4.54 |
ENSDART00000163137
|
elovl1a
|
ELOVL fatty acid elongase 1a |
| chr17_+_31346338 | 4.54 |
ENSDART00000062900
|
itpka
|
inositol-trisphosphate 3-kinase A |
| chr14_-_36523075 | 4.51 |
ENSDART00000109293
|
rnf130
|
ring finger protein 130 |
| chr9_-_14533551 | 4.50 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr17_+_15425559 | 4.48 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr16_-_7525980 | 4.41 |
ENSDART00000017445
|
prdm1a
|
PR domain containing 1a, with ZNF domain |
| chr17_-_30846176 | 4.36 |
ENSDART00000062811
|
yy1a
|
YY1 transcription factor a |
| chr5_-_23697487 | 4.28 |
ENSDART00000013309
|
sox19a
|
SRY (sex determining region Y)-box 19a |
| chr11_+_10925475 | 4.24 |
ENSDART00000064860
|
rbms1a
|
RNA binding motif, single stranded interacting protein 1a |
| chr22_+_37944019 | 4.22 |
|
|
|
| chr1_-_44888989 | 4.21 |
ENSDART00000135089
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr20_-_22036656 | 4.19 |
ENSDART00000152290
|
daam1b
|
dishevelled associated activator of morphogenesis 1b |
| chr25_-_207828 | 4.17 |
ENSDART00000169152
|
srpk2
|
SRSF protein kinase 2 |
| chr1_-_34921848 | 4.14 |
ENSDART00000142154
|
frem3
|
Fras1 related extracellular matrix 3 |
| chr9_-_22140954 | 4.10 |
ENSDART00000146528
|
lmo7a
|
LIM domain 7a |
| chr9_-_31467299 | 4.05 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr13_-_36996246 | 4.02 |
ENSDART00000133242
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr8_-_17480730 | 4.02 |
ENSDART00000100667
|
skia
|
v-ski avian sarcoma viral oncogene homolog a |
| chr20_-_44598129 | 4.00 |
ENSDART00000012229
|
fkbp1b
|
FK506 binding protein 1b |
| chr4_+_5733160 | 4.00 |
ENSDART00000110243
|
pou3f2a
|
POU class 3 homeobox 2a |
| chr9_-_32942783 | 3.99 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr10_+_3728747 | 3.98 |
|
|
|
| chr6_+_13613459 | 3.96 |
ENSDART00000104722
|
cdk5r2a
|
cyclin-dependent kinase 5, regulatory subunit 2a (p39) |
| chr22_+_659157 | 3.92 |
ENSDART00000149712
|
gpr37l1a
|
G protein-coupled receptor 37 like 1a |
| chr18_-_15404998 | 3.89 |
ENSDART00000031752
|
rfx4
|
regulatory factor X, 4 |
| chr4_-_6800721 | 3.88 |
ENSDART00000099467
|
ifrd1
|
interferon-related developmental regulator 1 |
| chr3_-_36339966 | 3.85 |
ENSDART00000176547
|
ppl
|
periplakin |
| chr17_+_18097490 | 3.84 |
ENSDART00000144894
|
bcl11ba
|
B-cell CLL/lymphoma 11Ba (zinc finger protein) |
| chr25_+_4490129 | 3.81 |
|
|
|
| chr16_+_17705704 | 3.78 |
|
|
|
| chr24_+_7607915 | 3.75 |
ENSDART00000124409
|
cavin1b
|
caveolae associated protein 1b |
| chr19_-_7501777 | 3.74 |
ENSDART00000081741
|
rab25a
|
RAB25, member RAS oncogene family a |
| chr16_-_39951493 | 3.73 |
|
|
|
| chr18_+_20504980 | 3.71 |
ENSDART00000060295
|
rapsn
|
receptor-associated protein of the synapse, 43kD |
| chr19_+_15538967 | 3.71 |
ENSDART00000171403
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr18_+_9213713 | 3.69 |
ENSDART00000127469
ENSDART00000101192 |
sema3d
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr24_-_21844170 | 3.59 |
ENSDART00000032963
|
apoob
|
apolipoprotein O, b |
| chr23_-_24240728 | 3.55 |
ENSDART00000113598
|
arhgef19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr16_+_16941228 | 3.49 |
ENSDART00000142155
|
myh14
|
myosin, heavy chain 14, non-muscle |
| chr5_-_65103664 | 3.49 |
ENSDART00000130888
|
notch1b
|
notch 1b |
| chr4_+_21485280 | 3.48 |
ENSDART00000066896
|
syt1a
|
synaptotagmin Ia |
| chr8_+_20125687 | 3.48 |
ENSDART00000124809
|
acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr2_-_30340646 | 3.47 |
ENSDART00000099078
|
jph1b
|
junctophilin 1b |
| chr16_+_21121428 | 3.44 |
|
|
|
| chr20_-_22036622 | 3.42 |
ENSDART00000152290
|
daam1b
|
dishevelled associated activator of morphogenesis 1b |
| chr21_+_17731439 | 3.41 |
ENSDART00000124173
|
rxraa
|
retinoid X receptor, alpha a |
| chr14_-_30363703 | 3.41 |
ENSDART00000134098
|
efemp2a
|
EGF containing fibulin-like extracellular matrix protein 2a |
| chr6_-_39008833 | 3.40 |
ENSDART00000150885
|
vdrb
|
vitamin D receptor b |
| chr18_-_15405161 | 3.39 |
ENSDART00000031752
|
rfx4
|
regulatory factor X, 4 |
| chr8_+_42992323 | 3.38 |
ENSDART00000048819
|
rassf2a
|
Ras association (RalGDS/AF-6) domain family member 2a |
| chr24_+_18804086 | 3.37 |
ENSDART00000106186
|
prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr21_-_36338514 | 3.35 |
ENSDART00000157826
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr13_+_19191645 | 3.31 |
ENSDART00000058036
|
emx2
|
empty spiracles homeobox 2 |
| chr9_-_7695437 | 3.29 |
ENSDART00000102715
|
tuba8l3
|
tubulin, alpha 8 like 3 |
| chr7_-_29811734 | 3.26 |
ENSDART00000075600
|
tspan3b
|
tetraspanin 3b |
| chr1_+_16681778 | 3.26 |
|
|
|
| chr2_-_10919978 | 3.25 |
ENSDART00000005944
|
rpl5a
|
ribosomal protein L5a |
| chr3_-_32190009 | 3.23 |
|
|
|
| chr15_-_20088439 | 3.23 |
ENSDART00000161379
|
auts2b
|
autism susceptibility candidate 2b |
| chr25_+_18467217 | 3.21 |
ENSDART00000170841
|
cav1
|
caveolin 1 |
| chr6_-_39009073 | 3.21 |
ENSDART00000150885
|
vdrb
|
vitamin D receptor b |
| KN149817v1_+_2207 | 3.21 |
|
|
|
| chr14_+_22889826 | 3.20 |
ENSDART00000170356
|
enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr1_-_11474337 | 3.19 |
ENSDART00000149913
|
b4galt1
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 |
| chr25_+_15551288 | 3.18 |
ENSDART00000137375
|
spon1b
|
spondin 1b |
| chr3_+_22412059 | 3.14 |
|
|
|
| chr10_-_23939618 | 3.12 |
|
|
|
| chr17_+_45834257 | 3.11 |
ENSDART00000035152
|
kif26ab
|
kinesin family member 26Ab |
| chr16_+_5470544 | 3.04 |
|
|
|
| chr10_-_32614936 | 2.98 |
ENSDART00000143301
|
mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr24_+_21200975 | 2.98 |
ENSDART00000126519
|
shisa2b
|
shisa family member 2b |
| chr7_+_40183905 | 2.97 |
ENSDART00000173916
|
ptprn2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr6_+_24717985 | 2.94 |
ENSDART00000165609
|
barhl2
|
BarH-like homeobox 2 |
| chr5_-_65103730 | 2.93 |
ENSDART00000169616
|
notch1b
|
notch 1b |
| chr23_-_23474703 | 2.91 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr3_+_17760847 | 2.90 |
ENSDART00000080946
|
ttc25
|
tetratricopeptide repeat domain 25 |
| chr13_-_31339870 | 2.88 |
ENSDART00000076574
|
rtn1a
|
reticulon 1a |
| chr23_-_7283514 | 2.87 |
ENSDART00000156369
|
CR589876.1
|
ENSDARG00000096997 |
| chr20_-_22584941 | 2.84 |
ENSDART00000063601
|
gsx2
|
GS homeobox 2 |
| chr24_-_16995194 | 2.82 |
ENSDART00000111079
|
CR376737.1
|
ENSDARG00000077318 |
| chr8_-_15144533 | 2.81 |
ENSDART00000138855
|
bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr1_-_22160662 | 2.79 |
ENSDART00000054386
|
qdprb1
|
quinoid dihydropteridine reductase b1 |
| chr3_-_38550961 | 2.79 |
ENSDART00000155042
|
mpp3a
|
membrane protein, palmitoylated 3a (MAGUK p55 subfamily member 3) |
| chr15_+_15479719 | 2.77 |
ENSDART00000127436
|
zgc:92630
|
zgc:92630 |
| chr25_+_15551474 | 2.77 |
ENSDART00000137375
|
spon1b
|
spondin 1b |
| chr10_+_10393377 | 2.76 |
ENSDART00000109432
|
cercam
|
cerebral endothelial cell adhesion molecule |
| chr15_+_36197093 | 2.73 |
ENSDART00000156697
|
col4a3
|
collagen, type IV, alpha 3 |
| chr21_-_36338704 | 2.71 |
ENSDART00000157826
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr15_-_36196994 | 2.71 |
ENSDART00000163061
|
col4a4
|
collagen, type IV, alpha 4 |
| chr20_-_14158114 | 2.69 |
ENSDART00000009549
|
rhag
|
Rh-associated glycoprotein |
| chr10_-_5135388 | 2.69 |
ENSDART00000108587
|
sec31a
|
SEC31 homolog A, COPII coat complex component |
| chr20_+_15119519 | 2.69 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr21_+_5887486 | 2.67 |
ENSDART00000048399
|
slc4a4b
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4b |
| chr23_-_3815492 | 2.64 |
ENSDART00000131536
|
hmga1a
|
high mobility group AT-hook 1a |
| chr6_-_34024063 | 2.64 |
ENSDART00000003701
|
tie1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr10_-_43924675 | 2.62 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr13_+_32013916 | 2.60 |
ENSDART00000020270
|
osr1
|
odd-skipped related transciption factor 1 |
| chr3_+_22962233 | 2.60 |
ENSDART00000142884
|
gngt2a
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2a |
| chr3_+_28822408 | 2.60 |
ENSDART00000133528
|
lgals2a
|
lectin, galactoside-binding, soluble, 2a |
| chr15_-_14616083 | 2.58 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr24_-_16994956 | 2.58 |
ENSDART00000111079
|
CR376737.1
|
ENSDARG00000077318 |
| chr12_-_43917936 | 2.57 |
ENSDART00000159423
|
foxi1
|
forkhead box i1 |
| chr8_-_15071283 | 2.56 |
|
|
|
| chr18_-_25002972 | 2.55 |
ENSDART00000163449
|
chd2
|
chromodomain helicase DNA binding protein 2 |
| chr9_-_2588401 | 2.54 |
ENSDART00000161018
|
sp9
|
sp9 transcription factor |
| chr2_+_16791661 | 2.52 |
ENSDART00000145107
|
agfg1b
|
ArfGAP with FG repeats 1b |
| chr13_-_33269297 | 2.52 |
|
|
|
| chr13_+_21695882 | 2.50 |
ENSDART00000165150
|
zswim8
|
zinc finger, SWIM-type containing 8 |
| chr7_-_46509291 | 2.50 |
ENSDART00000173891
|
tshz3b
|
teashirt zinc finger homeobox 3b |
| chr15_-_10345337 | 2.48 |
|
|
|
| chr18_-_46243753 | 2.46 |
ENSDART00000145999
|
prx
|
periaxin |
| chr1_-_40557436 | 2.45 |
ENSDART00000144424
|
rgs12b
|
regulator of G protein signaling 12b |
| chr19_+_17941678 | 2.45 |
ENSDART00000124597
|
nr1d2b
|
nuclear receptor subfamily 1, group D, member 2b |
| chr9_+_4188263 | 2.45 |
ENSDART00000164543
|
kalrna
|
kalirin RhoGEF kinase a |
| chr5_+_51392688 | 2.44 |
ENSDART00000143276
|
setbp1
|
SET binding protein 1 |
| chr10_+_5693933 | 2.44 |
ENSDART00000159769
|
pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr15_-_9443120 | 2.43 |
ENSDART00000177158
|
sacs
|
sacsin molecular chaperone |
| chr9_-_21257082 | 2.43 |
ENSDART00000124533
|
tbx15
|
T-box 15 |
| chr4_+_22020246 | 2.42 |
ENSDART00000112035
ENSDART00000127664 |
myf5
|
myogenic factor 5 |
| chr9_-_28464236 | 2.42 |
ENSDART00000146932
|
creb1b
|
cAMP responsive element binding protein 1b |
| chr3_+_32360862 | 2.40 |
ENSDART00000151329
|
trpm4a
|
transient receptor potential cation channel, subfamily M, member 4a |
| chr7_-_58649991 | 2.37 |
ENSDART00000167076
|
mrc1a
|
mannose receptor, C type 1a |
| chr11_+_6079418 | 2.36 |
ENSDART00000158675
|
ushbp1
|
Usher syndrome 1C binding protein 1 |
| chr16_-_38604326 | 2.34 |
ENSDART00000109124
|
rspo2
|
R-spondin 2 |
| chr2_-_32468715 | 2.32 |
|
|
|
| chr15_-_32738134 | 2.31 |
ENSDART00000164670
|
frem2b
|
Fras1 related extracellular matrix protein 2b |
| chr3_-_30554400 | 2.31 |
ENSDART00000151097
|
si:ch211-51c14.1
|
si:ch211-51c14.1 |
| chr20_-_21773202 | 2.30 |
ENSDART00000133286
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr13_-_5808515 | 2.29 |
ENSDART00000165625
|
spred2b
|
sprouty-related, EVH1 domain containing 2b |
| chr5_-_20441931 | 2.26 |
|
|
|
| chr8_-_6875110 | 2.25 |
ENSDART00000014915
|
asb6
|
ankyrin repeat and SOCS box containing 6 |
| chr8_-_3253562 | 2.25 |
ENSDART00000035965
|
fut9b
|
fucosyltransferase 9b |
| chr8_-_3974357 | 2.22 |
ENSDART00000163754
ENSDART00000169474 |
mtmr3
|
myotubularin related protein 3 |
| chr4_-_11595364 | 2.21 |
ENSDART00000002682
|
net1
|
neuroepithelial cell transforming 1 |
| chr4_-_4825948 | 2.21 |
ENSDART00000141539
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr19_-_35828807 | 2.20 |
|
|
|
| chr11_-_25017693 | 2.20 |
|
|
|
| chr2_+_23038657 | 2.19 |
ENSDART00000089012
|
kif1ab
|
kinesin family member 1Ab |
| chr1_+_48708504 | 2.19 |
ENSDART00000008468
|
msx1b
|
muscle segment homeobox 1b |
| chr20_+_14979154 | 2.18 |
ENSDART00000118157
|
dre-mir-199-1
|
dre-mir-199-1 |
| chr18_+_17594381 | 2.16 |
ENSDART00000010998
|
slc12a3
|
solute carrier family 12 (sodium/chloride transporter), member 3 |
| chr24_+_37692365 | 2.14 |
|
|
|
| chr13_-_18704185 | 2.14 |
ENSDART00000146795
|
lzts2a
|
leucine zipper, putative tumor suppressor 2a |
| chr16_+_17706003 | 2.12 |
|
|
|
| chr5_+_22465722 | 2.10 |
ENSDART00000145477
ENSDART00000089992 |
hmgn7
|
high mobility group nucleosomal binding domain 7 |
| chr12_+_3189489 | 2.09 |
ENSDART00000010569
|
g6pc3
|
glucose 6 phosphatase, catalytic, 3 |
| chr20_+_38298893 | 2.08 |
ENSDART00000022694
|
ehd3
|
EH-domain containing 3 |
| chr8_+_47352102 | 2.08 |
|
|
|
| chr20_+_18310624 | 2.06 |
ENSDART00000111063
|
kctd1
|
potassium channel tetramerization domain containing 1 |
| chr5_+_23585492 | 2.05 |
ENSDART00000135083
|
tp53
|
tumor protein p53 |
| chr24_-_21324086 | 2.05 |
|
|
|
| chr24_+_34227202 | 2.02 |
ENSDART00000105572
|
gbx1
|
gastrulation brain homeobox 1 |
| chr20_-_40217185 | 2.02 |
|
|
|
| chr19_+_3925188 | 2.01 |
|
|
|
| chr21_-_24552672 | 1.99 |
|
|
|
| chr17_+_24428093 | 1.98 |
ENSDART00000131417
|
ugp2b
|
UDP-glucose pyrophosphorylase 2b |
| chr22_-_20360142 | 1.97 |
ENSDART00000089015
|
zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr21_+_25757285 | 1.96 |
ENSDART00000101217
|
ENSDARG00000069505
|
ENSDARG00000069505 |
| chr17_+_44666606 | 1.94 |
ENSDART00000155096
|
tmem63c
|
transmembrane protein 63C |
| chr21_+_25150600 | 1.94 |
ENSDART00000101147
|
si:dkey-183i3.5
|
si:dkey-183i3.5 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.4 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 3.1 | 9.2 | GO:0098743 | cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 2.9 | 8.8 | GO:0061323 | cell proliferation involved in heart morphogenesis(GO:0061323) regulation of cell proliferation involved in heart morphogenesis(GO:2000136) |
| 2.7 | 8.2 | GO:0032369 | negative regulation of lipid transport(GO:0032369) |
| 2.6 | 10.5 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) pharyngeal muscle development(GO:0043282) |
| 2.4 | 7.1 | GO:0050810 | regulation of steroid biosynthetic process(GO:0050810) |
| 1.8 | 7.3 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 1.8 | 5.3 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 1.3 | 2.7 | GO:0019755 | one-carbon compound transport(GO:0019755) |
| 1.3 | 19.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 1.3 | 4.0 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 1.2 | 3.7 | GO:0007508 | larval development(GO:0002164) larval heart development(GO:0007508) |
| 1.2 | 4.6 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 1.1 | 4.6 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 1.1 | 3.2 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 1.0 | 3.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 1.0 | 9.6 | GO:0006956 | complement activation(GO:0006956) |
| 0.9 | 2.6 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.8 | 2.5 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.8 | 6.4 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.8 | 2.4 | GO:0002706 | regulation of lymphocyte mediated immunity(GO:0002706) |
| 0.8 | 4.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.8 | 4.7 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.8 | 4.5 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.7 | 5.2 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.7 | 3.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.6 | 2.6 | GO:0035776 | pronephric proximal tubule development(GO:0035776) |
| 0.6 | 5.2 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.6 | 4.4 | GO:0051155 | positive regulation of striated muscle cell differentiation(GO:0051155) |
| 0.6 | 1.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.6 | 2.3 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.5 | 1.6 | GO:1990092 | calcium-dependent self proteolysis(GO:1990092) |
| 0.5 | 1.6 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.5 | 1.5 | GO:0006014 | D-ribose metabolic process(GO:0006014) pentose catabolic process(GO:0019323) |
| 0.4 | 1.3 | GO:0097104 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.4 | 2.5 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.4 | 4.0 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.4 | 1.9 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.4 | 2.6 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.4 | 3.3 | GO:0060114 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.4 | 4.2 | GO:0090309 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.3 | 5.7 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.3 | 3.5 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.3 | 5.2 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.3 | 2.4 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.3 | 1.2 | GO:0045168 | cell-cell signaling involved in cell fate commitment(GO:0045168) |
| 0.3 | 1.8 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.3 | 4.2 | GO:0001952 | regulation of cell-matrix adhesion(GO:0001952) |
| 0.3 | 7.4 | GO:0060119 | inner ear receptor cell development(GO:0060119) |
| 0.3 | 3.8 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.3 | 2.8 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.3 | 2.2 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.3 | 0.8 | GO:0051963 | regulation of synapse assembly(GO:0051963) positive regulation of synapse assembly(GO:0051965) |
| 0.3 | 2.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.3 | 5.1 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.3 | 2.0 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.2 | 1.2 | GO:2000651 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.2 | 3.4 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.2 | 1.4 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.2 | 2.6 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.2 | 1.2 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.2 | 4.5 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 | 0.9 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.2 | 2.1 | GO:0001964 | startle response(GO:0001964) |
| 0.2 | 1.1 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.2 | 2.9 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.2 | 2.3 | GO:0033336 | caudal fin development(GO:0033336) |
| 0.2 | 4.9 | GO:0039020 | pronephric nephron tubule development(GO:0039020) |
| 0.2 | 4.5 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 4.3 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.2 | 2.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 6.9 | GO:0045104 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.2 | 3.6 | GO:0042407 | cristae formation(GO:0042407) |
| 0.2 | 2.2 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.2 | 1.5 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.2 | 3.4 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.2 | 3.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.2 | 2.5 | GO:1902667 | regulation of axon guidance(GO:1902667) |
| 0.2 | 3.9 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.2 | 0.7 | GO:1901998 | toxin transport(GO:1901998) |
| 0.2 | 0.9 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.2 | 1.5 | GO:0035437 | maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.2 | 0.8 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.2 | 0.9 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.2 | 3.8 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.1 | 1.6 | GO:0006959 | humoral immune response(GO:0006959) |
| 0.1 | 2.1 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.1 | 3.7 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.1 | 3.5 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.1 | 4.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.7 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.1 | 0.9 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.1 | 0.8 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.1 | 1.2 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.1 | 0.3 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 0.4 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 1.6 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 1.3 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.1 | 6.7 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 1.5 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.1 | 0.7 | GO:0010526 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.1 | 11.7 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.1 | 0.9 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.1 | 0.8 | GO:0045117 | azole transport(GO:0045117) |
| 0.1 | 0.6 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 3.7 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 2.2 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.1 | 0.4 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.1 | 2.6 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 7.7 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.1 | 4.3 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.1 | 1.9 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.1 | 2.2 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.1 | 1.5 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.1 | 1.0 | GO:0045738 | negative regulation of DNA repair(GO:0045738) negative regulation of double-strand break repair(GO:2000780) |
| 0.1 | 1.2 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 1.8 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 1.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.7 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 1.4 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.1 | 1.1 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 1.1 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 0.2 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 0.8 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 2.7 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.1 | 5.6 | GO:0030155 | regulation of cell adhesion(GO:0030155) |
| 0.1 | 2.4 | GO:0009755 | hormone-mediated signaling pathway(GO:0009755) |
| 0.1 | 8.1 | GO:0060026 | convergent extension(GO:0060026) |
| 0.1 | 0.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 3.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.2 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.9 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.8 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 2.9 | GO:0051961 | negative regulation of neurogenesis(GO:0050768) negative regulation of nervous system development(GO:0051961) |
| 0.0 | 1.9 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 2.4 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 0.8 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.5 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.3 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 5.4 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.7 | GO:0008217 | regulation of blood pressure(GO:0008217) |
| 0.0 | 2.4 | GO:0001708 | cell fate specification(GO:0001708) |
| 0.0 | 1.5 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 1.3 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.6 | GO:0043489 | RNA stabilization(GO:0043489) |
| 0.0 | 0.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 1.3 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 0.6 | GO:0046460 | neutral lipid biosynthetic process(GO:0046460) acylglycerol biosynthetic process(GO:0046463) |
| 0.0 | 0.4 | GO:0060840 | artery development(GO:0060840) |
| 0.0 | 3.2 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.3 | GO:0046113 | nucleobase catabolic process(GO:0046113) |
| 0.0 | 3.5 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 0.3 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.8 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 0.9 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.2 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 1.4 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 6.9 | GO:0007420 | brain development(GO:0007420) |
| 0.0 | 0.1 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.9 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.1 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 1.8 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 1.4 | GO:0070507 | regulation of microtubule-based process(GO:0032886) regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 2.1 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
| 0.0 | 0.2 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 1.1 | GO:0050767 | regulation of neurogenesis(GO:0050767) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 1.0 | 4.0 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.9 | 8.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.8 | 9.2 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.6 | 1.8 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.5 | 12.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.5 | 2.6 | GO:0043034 | costamere(GO:0043034) |
| 0.5 | 2.5 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.4 | 1.9 | GO:0016234 | inclusion body(GO:0016234) |
| 0.3 | 3.7 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.3 | 9.8 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.3 | 3.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.3 | 1.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.3 | 3.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.3 | 1.9 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.2 | 2.9 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 2.6 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.2 | 13.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 2.4 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.2 | 1.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.2 | 8.5 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 0.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 18.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 8.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.7 | GO:1904949 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.1 | 4.4 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 1.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 3.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 7.4 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.1 | 36.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 0.7 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 15.8 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 3.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 4.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 1.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 2.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 0.7 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.1 | 0.8 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 3.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 2.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 2.4 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.6 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 3.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.0 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 5.6 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 4.3 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 2.5 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 1.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.6 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 2.2 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 1.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 2.1 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 1.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.2 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 4.3 | GO:0015630 | microtubule cytoskeleton(GO:0015630) |
| 0.0 | 0.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.5 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 1.5 | 4.5 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 1.3 | 5.2 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 1.2 | 8.1 | GO:0070888 | E-box binding(GO:0070888) |
| 1.1 | 12.3 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.7 | 2.8 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.7 | 3.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.7 | 3.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.6 | 2.6 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.6 | 3.8 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.6 | 2.4 | GO:0016842 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) amidine-lyase activity(GO:0016842) |
| 0.5 | 1.6 | GO:0070548 | glutamine-fructose-6-phosphate transaminase (isomerizing) activity(GO:0004360) L-glutamine aminotransferase activity(GO:0070548) |
| 0.5 | 3.7 | GO:0043495 | protein anchor(GO:0043495) |
| 0.5 | 4.5 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.5 | 5.7 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.5 | 8.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.4 | 7.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.4 | 4.0 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.4 | 4.3 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.4 | 1.2 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.4 | 4.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.4 | 22.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.4 | 2.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.4 | 1.9 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.3 | 1.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.3 | 5.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.3 | 1.8 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.3 | 2.7 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.3 | 0.9 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.3 | 3.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.3 | 3.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.3 | 15.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.3 | 3.5 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.2 | 1.2 | GO:0017081 | chloride channel regulator activity(GO:0017081) chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.2 | 1.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.2 | 1.4 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.2 | 0.9 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.2 | 1.5 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.2 | 2.2 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.2 | 2.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 4.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 9.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.2 | 6.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 7.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.2 | 3.6 | GO:0016411 | acylglycerol O-acyltransferase activity(GO:0016411) |
| 0.2 | 4.5 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 2.9 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.2 | 1.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.9 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.1 | 1.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.8 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 0.8 | GO:0045118 | azole transporter activity(GO:0045118) vitamin transmembrane transporter activity(GO:0090482) azole transmembrane transporter activity(GO:1901474) |
| 0.1 | 5.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 4.4 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 4.7 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 1.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.3 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.1 | 9.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.4 | GO:0051717 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.1 | 0.5 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 3.9 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.1 | 1.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.0 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 7.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 16.5 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.1 | 6.7 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 0.3 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) molybdenum ion binding(GO:0030151) |
| 0.1 | 1.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 5.3 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.1 | 1.2 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 1.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 4.0 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 2.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 2.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 2.6 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 3.3 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.9 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 7.0 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 1.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.2 | GO:0008988 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) |
| 0.0 | 59.6 | GO:1990837 | sequence-specific double-stranded DNA binding(GO:1990837) |
| 0.0 | 0.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.4 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 9.9 | GO:0038023 | signaling receptor activity(GO:0038023) |
| 0.0 | 1.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 3.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.3 | GO:0061733 | peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 10.6 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.8 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 1.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 1.8 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 5.7 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 1.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 2.9 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 1.3 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.0 | 2.3 | GO:0005543 | phospholipid binding(GO:0005543) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 6.4 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.6 | 6.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.6 | 4.6 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.4 | 19.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.4 | 8.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 8.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 2.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 3.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 4.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 11.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 4.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 1.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 1.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 2.1 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 2.4 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 2.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.7 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.7 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 1.8 | 8.8 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.9 | 22.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.6 | 4.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.6 | 12.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.5 | 10.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.4 | 6.8 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.3 | 2.7 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.3 | 3.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.3 | 0.8 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.2 | 4.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 3.8 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 1.5 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.1 | 1.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 3.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 0.9 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 0.7 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 0.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 0.5 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 2.1 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 0.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 0.7 | REACTOME SIGNALING BY FGFR1 MUTANTS | Genes involved in Signaling by FGFR1 mutants |
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 1.0 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |