Project
DANIO-CODE
Navigation
Downloads
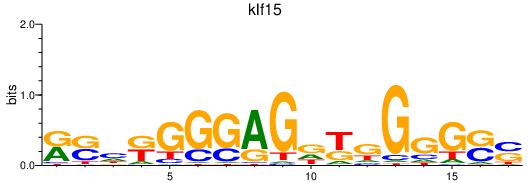
Results for klf15
Z-value: 0.79
Transcription factors associated with klf15
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
klf15
|
ENSDARG00000091127 | Kruppel-like factor 15 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| klf15 | dr10_dc_chr23_-_35096962_35097011 | -0.63 | 8.9e-03 | Click! |
Activity profile of klf15 motif
Sorted Z-values of klf15 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of klf15
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_21563703 | 2.06 |
ENSDART00000045574
|
shisa2a
|
shisa family member 2a |
| chr21_-_13593659 | 1.87 |
ENSDART00000065817
|
pou5f3
|
POU domain, class 5, transcription factor 3 |
| chr14_+_34154895 | 1.61 |
ENSDART00000147756
|
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr5_+_26925392 | 1.47 |
ENSDART00000051491
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr24_-_4418454 | 1.44 |
ENSDART00000066835
|
fzd8a
|
frizzled class receptor 8a |
| chr5_+_26925238 | 1.31 |
ENSDART00000051491
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr3_-_18653909 | 1.27 |
|
|
|
| KN149861v1_-_5655 | 1.25 |
|
|
|
| chr22_-_20377970 | 1.18 |
ENSDART00000010048
|
map2k2a
|
mitogen-activated protein kinase kinase 2a |
| KN149861v1_-_6089 | 1.17 |
|
|
|
| chr5_+_26925663 | 1.00 |
ENSDART00000051491
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr16_+_9822930 | 0.98 |
ENSDART00000164103
|
ecm1b
|
extracellular matrix protein 1b |
| chr8_+_17479651 | 0.89 |
|
|
|
| chr23_+_31318844 | 0.82 |
ENSDART00000146510
|
irak1bp1
|
interleukin-1 receptor-associated kinase 1 binding protein 1 |
| chr15_-_18087085 | 0.81 |
ENSDART00000155194
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr5_-_29480173 | 0.80 |
ENSDART00000140049
|
bco2a
|
beta-carotene oxygenase 2a |
| chr19_+_14132374 | 0.79 |
ENSDART00000164696
|
tmem222b
|
transmembrane protein 222b |
| chr4_-_6443699 | 0.77 |
|
|
|
| chr25_+_29631947 | 0.77 |
ENSDART00000154849
|
ENSDARG00000029431
|
ENSDARG00000029431 |
| chr19_+_14490203 | 0.75 |
ENSDART00000164386
|
arid1ab
|
AT rich interactive domain 1Ab (SWI-like) |
| chr18_-_26876598 | 0.73 |
|
|
|
| chr11_+_6012910 | 0.72 |
ENSDART00000161001
|
gtpbp3
|
GTP binding protein 3 |
| chr19_+_14132473 | 0.71 |
ENSDART00000164696
|
tmem222b
|
transmembrane protein 222b |
| chr3_-_22240424 | 0.70 |
|
|
|
| chr14_+_46609215 | 0.69 |
ENSDART00000110429
|
zgc:194285
|
zgc:194285 |
| chr5_+_26925957 | 0.69 |
|
|
|
| chr5_-_56641212 | 0.67 |
ENSDART00000050957
|
fer
|
fer (fps/fes related) tyrosine kinase |
| chr18_+_46153484 | 0.61 |
ENSDART00000015034
|
blvrb
|
biliverdin reductase B |
| chr16_+_9823111 | 0.56 |
ENSDART00000164103
|
ecm1b
|
extracellular matrix protein 1b |
| chr5_-_56641001 | 0.55 |
ENSDART00000149855
|
fer
|
fer (fps/fes related) tyrosine kinase |
| chr21_+_15505169 | 0.53 |
ENSDART00000011318
|
smarcb1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1b |
| chr13_+_31586024 | 0.53 |
ENSDART00000034745
|
prkcha
|
protein kinase C, eta, a |
| chr2_-_56848527 | 0.52 |
ENSDART00000164330
|
CU634008.1
|
ENSDARG00000100430 |
| chr12_+_10405904 | 0.52 |
ENSDART00000029133
|
snu13b
|
SNU13 homolog, small nuclear ribonucleoprotein b (U4/U6.U5) |
| chr5_-_56641061 | 0.49 |
ENSDART00000050957
|
fer
|
fer (fps/fes related) tyrosine kinase |
| chr9_-_18808049 | 0.48 |
|
|
|
| chr7_-_71092598 | 0.47 |
ENSDART00000149682
|
dhx15
|
DEAH (Asp-Glu-Ala-His) box helicase 15 |
| chr8_-_52952141 | 0.47 |
ENSDART00000168185
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr19_-_17039269 | 0.47 |
|
|
|
| chr10_-_25695574 | 0.47 |
ENSDART00000110751
|
tiam1a
|
T-cell lymphoma invasion and metastasis 1a |
| chr17_+_15527525 | 0.47 |
ENSDART00000159426
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr22_-_5888816 | 0.47 |
ENSDART00000141373
|
si:rp71-36a1.1
|
si:rp71-36a1.1 |
| chr17_-_2544896 | 0.45 |
ENSDART00000109582
|
spata7
|
spermatogenesis associated 7 |
| chr16_+_9822879 | 0.43 |
ENSDART00000164103
|
ecm1b
|
extracellular matrix protein 1b |
| chr5_+_21563654 | 0.43 |
ENSDART00000045574
|
shisa2a
|
shisa family member 2a |
| chr12_+_23300259 | 0.43 |
|
|
|
| chr7_+_19300487 | 0.43 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr6_-_29386437 | 0.42 |
ENSDART00000168905
|
tmem131
|
transmembrane protein 131 |
| chr9_-_18807868 | 0.40 |
|
|
|
| chr3_+_35310412 | 0.39 |
ENSDART00000176700
|
BX511029.1
|
ENSDARG00000107881 |
| chr22_-_20378023 | 0.38 |
ENSDART00000010048
|
map2k2a
|
mitogen-activated protein kinase kinase 2a |
| chr6_+_36843778 | 0.38 |
ENSDART00000104157
|
tmem45a
|
transmembrane protein 45a |
| chr8_+_8908955 | 0.37 |
ENSDART00000131215
|
slc35a2
|
solute carrier family 35 (UDP-galactose transporter), member 2 |
| chr8_-_40231417 | 0.34 |
ENSDART00000162020
|
kdm2ba
|
lysine (K)-specific demethylase 2Ba |
| chr21_+_7627327 | 0.32 |
ENSDART00000172174
|
agpat2
|
1-acylglycerol-3-phosphate O-acyltransferase 2 (lysophosphatidic acid acyltransferase, beta) |
| chr9_+_23118780 | 0.32 |
ENSDART00000061299
|
tsn
|
translin |
| chr7_+_19300351 | 0.32 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr16_-_39527598 | 0.31 |
ENSDART00000102525
|
stt3b
|
STT3B, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr16_+_51598269 | 0.31 |
ENSDART00000169443
|
SLC9A1
|
solute carrier family 9 member A1 |
| KN149861v1_-_5543 | 0.31 |
|
|
|
| chr8_-_52952226 | 0.31 |
ENSDART00000168185
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr6_-_10574082 | 0.30 |
ENSDART00000151661
|
wipf1b
|
WAS/WASL interacting protein family, member 1b |
| chr11_-_40239946 | 0.30 |
ENSDART00000165394
|
si:dkeyp-61b2.1
|
si:dkeyp-61b2.1 |
| chr11_+_33154781 | 0.29 |
|
|
|
| chr1_-_51862897 | 0.28 |
ENSDART00000136469
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr11_+_11735899 | 0.28 |
ENSDART00000003891
|
jupa
|
junction plakoglobin a |
| chr2_-_7374447 | 0.28 |
|
|
|
| chr4_-_195110 | 0.27 |
ENSDART00000167885
|
lrp6
|
low density lipoprotein receptor-related protein 6 |
| chr5_-_34664282 | 0.25 |
ENSDART00000114981
ENSDART00000051279 |
ptcd2
|
pentatricopeptide repeat domain 2 |
| chr11_-_29891067 | 0.24 |
ENSDART00000172106
|
scml2
|
sex comb on midleg-like 2 (Drosophila) |
| chr3_-_25239180 | 0.24 |
ENSDART00000055491
|
smurf2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr2_+_25212281 | 0.23 |
ENSDART00000078838
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr12_+_10078043 | 0.22 |
ENSDART00000152369
|
si:dkeyp-118b1.2
|
si:dkeyp-118b1.2 |
| chr3_-_51724440 | 0.22 |
|
|
|
| chr12_-_14172962 | 0.22 |
ENSDART00000158399
|
avl9
|
AVL9 homolog (S. cerevisiase) |
| chr3_+_32293441 | 0.21 |
ENSDART00000156464
|
prr12b
|
proline rich 12b |
| chr2_-_44330306 | 0.21 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
| chr7_+_23224924 | 0.20 |
ENSDART00000142401
|
zgc:109889
|
zgc:109889 |
| chr18_-_26732762 | 0.20 |
ENSDART00000142043
|
malt3
|
MALT paracaspase 3 |
| chr22_+_26840517 | 0.18 |
ENSDART00000158756
|
crebbpa
|
CREB binding protein a |
| chr7_+_23224800 | 0.17 |
ENSDART00000115299
ENSDART00000101423 |
zgc:109889
|
zgc:109889 |
| chr9_-_32295031 | 0.15 |
ENSDART00000132448
|
BX530407.4
|
ENSDARG00000093950 |
| chr4_-_194901 | 0.15 |
ENSDART00000166440
ENSDART00000168093 |
lrp6
|
low density lipoprotein receptor-related protein 6 |
| chr17_-_2544927 | 0.14 |
ENSDART00000109582
|
spata7
|
spermatogenesis associated 7 |
| chr17_-_52735527 | 0.13 |
|
|
|
| chr17_+_8166308 | 0.12 |
ENSDART00000172443
|
cdc42bpaa
|
CDC42 binding protein kinase alpha (DMPK-like) a |
| chr25_+_65335 | 0.11 |
ENSDART00000170892
ENSDART00000158574 ENSDART00000008663 |
adam10b
|
ADAM metallopeptidase domain 10b |
| chr22_-_3134699 | 0.11 |
ENSDART00000159580
|
lmnb2
|
lamin B2 |
| chr13_+_28601627 | 0.10 |
ENSDART00000015773
|
ldb1a
|
LIM domain binding 1a |
| chr5_-_14147868 | 0.08 |
ENSDART00000113037
|
sema4c
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr4_-_195193 | 0.08 |
ENSDART00000167885
|
lrp6
|
low density lipoprotein receptor-related protein 6 |
| chr16_+_25845432 | 0.06 |
|
|
|
| chr23_+_41295092 | 0.04 |
ENSDART00000109567
|
nhsa
|
Nance-Horan syndrome a (congenital cataracts and dental anomalies) |
| chr22_-_95158 | 0.04 |
|
|
|
| chr14_-_23501428 | 0.03 |
|
|
|
| chr7_-_68033162 | 0.02 |
|
|
|
| chr18_-_14797284 | 0.02 |
|
|
|
| chr3_+_23573053 | 0.02 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr5_-_13844232 | 0.00 |
|
|
|
| chr17_+_31346473 | 0.00 |
ENSDART00000062900
|
itpka
|
inositol-trisphosphate 3-kinase A |
| chr2_-_44330348 | 0.00 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.4 | 1.6 | GO:0021512 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.3 | 1.7 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.3 | 1.9 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.3 | 0.8 | GO:0016121 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.2 | 0.5 | GO:2001284 | epithelial cell morphogenesis involved in gastrulation(GO:0003381) BMP secretion(GO:0038055) positive regulation of BMP secretion(GO:1900144) regulation of BMP secretion(GO:2001284) |
| 0.1 | 3.8 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 2.0 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.1 | 0.2 | GO:0048790 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.6 | GO:0097499 | protein localization to nonmotile primary cilium(GO:0097499) |
| 0.0 | 1.6 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.3 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.7 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.3 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.4 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.4 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.3 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.5 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
| 0.0 | 0.5 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.8 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.7 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.5 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 0.8 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.1 | 1.3 | GO:0035060 | brahma complex(GO:0035060) |
| 0.1 | 0.8 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.5 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.7 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.6 | GO:0005930 | axoneme(GO:0005930) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 5.7 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 1.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.3 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 1.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.4 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.5 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 1.9 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.7 | GO:0031491 | nucleosome binding(GO:0031491) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 2.1 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.7 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |