Project
DANIO-CODE
Navigation
Downloads
Results for klf3+klf8
Z-value: 0.71
Transcription factors associated with klf3+klf8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
klf3
|
ENSDARG00000015495 | Kruppel-like factor 3 (basic) |
|
klf8
|
ENSDARG00000060661 | Kruppel-like factor 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| klf3 | dr10_dc_chr1_-_17922028_17922172 | 0.78 | 3.4e-04 | Click! |
| klf8 | dr10_dc_chr21_+_37985816_37985880 | 0.68 | 3.6e-03 | Click! |
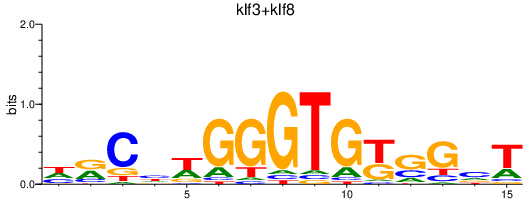
Activity profile of klf3+klf8 motif
Sorted Z-values of klf3+klf8 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of klf3+klf8
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_29153215 | 3.02 |
ENSDART00000156799
|
si:ch211-137a8.4
|
si:ch211-137a8.4 |
| chr16_+_24063849 | 2.39 |
ENSDART00000135084
|
apoa2
|
apolipoprotein A-II |
| chr14_+_13147115 | 2.32 |
ENSDART00000054849
|
pls3
|
plastin 3 (T isoform) |
| chr20_+_17840364 | 2.29 |
ENSDART00000024627
|
cdh2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr15_+_46114986 | 1.92 |
ENSDART00000156366
|
si:ch1073-340i21.3
|
si:ch1073-340i21.3 |
| chr3_-_32795866 | 1.80 |
ENSDART00000140117
|
aoc2
|
amine oxidase, copper containing 2 |
| chr18_-_45750 | 1.58 |
ENSDART00000148821
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr2_-_35584440 | 1.57 |
ENSDART00000125298
|
tnw
|
tenascin W |
| chr16_-_17148579 | 1.34 |
ENSDART00000138715
|
plekhg6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr18_+_5456221 | 1.17 |
|
|
|
| chr1_-_48790379 | 1.06 |
ENSDART00000150842
|
USMG5
|
up-regulated during skeletal muscle growth 5 homolog (mouse) |
| chr18_-_45892 | 1.04 |
ENSDART00000052641
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr10_+_4874841 | 1.01 |
ENSDART00000165942
|
palm2
|
paralemmin 2 |
| chr1_-_44239584 | 0.97 |
ENSDART00000137216
|
tmem176
|
transmembrane protein 176 |
| chr21_+_17505748 | 0.97 |
ENSDART00000163238
|
stom
|
stomatin |
| chr6_-_23193752 | 0.92 |
ENSDART00000159749
|
usp43a
|
ubiquitin specific peptidase 43a |
| chr5_-_68270398 | 0.89 |
ENSDART00000161561
|
ENSDARG00000059093
|
ENSDARG00000059093 |
| chr13_+_1308683 | 0.87 |
ENSDART00000159047
|
si:ch211-165e15.1
|
si:ch211-165e15.1 |
| chr22_-_11408859 | 0.86 |
ENSDART00000007649
|
mid1ip1b
|
MID1 interacting protein 1b |
| chr23_-_1008307 | 0.86 |
ENSDART00000110588
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr18_+_7350133 | 0.86 |
ENSDART00000172404
|
cers3b
|
ceramide synthase 3b |
| chr1_+_54460288 | 0.86 |
ENSDART00000138070
ENSDART00000150510 |
mb
|
myoglobin |
| chr8_-_21062262 | 0.85 |
|
|
|
| KN149978v1_-_10669 | 0.78 |
ENSDART00000175723
|
CABZ01068178.1
|
ENSDARG00000105971 |
| chr23_-_7889744 | 0.74 |
ENSDART00000164117
|
myt1b
|
myelin transcription factor 1b |
| chr21_+_13063614 | 0.73 |
|
|
|
| chr25_-_36886647 | 0.72 |
ENSDART00000160688
|
psma1
|
proteasome subunit alpha 1 |
| chr24_-_40968409 | 0.72 |
ENSDART00000169315
ENSDART00000171543 |
smyhc1
|
slow myosin heavy chain 1 |
| chr5_-_71394207 | 0.70 |
ENSDART00000050971
|
rab14l
|
RAB14, member RAS oncogene family, like |
| chr15_-_9443120 | 0.66 |
ENSDART00000177158
|
sacs
|
sacsin molecular chaperone |
| chr12_-_48278273 | 0.66 |
|
|
|
| chr6_+_170792 | 0.66 |
|
|
|
| chr2_-_1753058 | 0.65 |
ENSDART00000126566
|
slc22a23
|
solute carrier family 22, member 23 |
| chr21_+_44833070 | 0.59 |
ENSDART00000065083
ENSDART00000168217 |
fstl4
|
follistatin-like 4 |
| chr24_-_14068113 | 0.59 |
ENSDART00000130825
|
xkr9
|
XK, Kell blood group complex subunit-related family, member 9 |
| chr18_+_9524247 | 0.59 |
ENSDART00000053125
|
sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr7_+_36267647 | 0.57 |
ENSDART00000173653
|
chd9
|
chromodomain helicase DNA binding protein 9 |
| chr12_+_34662699 | 0.56 |
ENSDART00000153097
|
slc38a10
|
solute carrier family 38, member 10 |
| chr19_-_35642440 | 0.55 |
ENSDART00000167853
ENSDART00000054274 |
macf1a
|
microtubule-actin crosslinking factor 1a |
| chr1_+_44239727 | 0.54 |
ENSDART00000141145
|
si:dkey-9i23.16
|
si:dkey-9i23.16 |
| chr15_-_47264693 | 0.52 |
|
|
|
| chr10_+_34372205 | 0.48 |
ENSDART00000110121
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr6_-_39346614 | 0.44 |
ENSDART00000104074
|
zgc:158846
|
zgc:158846 |
| chr10_+_36096599 | 0.44 |
|
|
|
| chr7_-_6215188 | 0.39 |
ENSDART00000159542
|
zgc:112234
|
zgc:112234 |
| chr8_-_30233570 | 0.39 |
ENSDART00000143809
|
zgc:162939
|
zgc:162939 |
| chr5_-_63531729 | 0.39 |
ENSDART00000171711
|
gpsm1a
|
G protein signaling modulator 1a |
| chr16_-_12375204 | 0.39 |
ENSDART00000113345
|
leng8
|
leukocyte receptor cluster (LRC) member 8 |
| chr2_-_17721575 | 0.36 |
ENSDART00000141188
ENSDART00000100201 |
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr6_-_60088120 | 0.35 |
|
|
|
| chr21_+_45622590 | 0.35 |
ENSDART00000162422
|
sar1b
|
secretion associated, Ras related GTPase 1B |
| chr1_+_44239682 | 0.34 |
ENSDART00000141145
|
si:dkey-9i23.16
|
si:dkey-9i23.16 |
| chr2_-_49060868 | 0.34 |
|
|
|
| chr6_-_23193873 | 0.33 |
ENSDART00000159749
|
usp43a
|
ubiquitin specific peptidase 43a |
| chr1_+_48790583 | 0.32 |
ENSDART00000124833
|
pdcd11
|
programmed cell death 11 |
| chr21_+_11151424 | 0.31 |
ENSDART00000163432
|
arid6
|
AT-rich interaction domain 6 |
| chr15_-_47264619 | 0.29 |
|
|
|
| chr2_-_54909304 | 0.28 |
ENSDART00000100103
|
acss2l
|
acyl-CoA synthetase short-chain family member 2 like |
| chr2_-_6351592 | 0.27 |
|
|
|
| chr20_+_50091972 | 0.25 |
ENSDART00000097865
|
ints9
|
integrator complex subunit 9 |
| chr18_+_54352 | 0.25 |
ENSDART00000097163
|
SLC27A2 (1 of many)
|
solute carrier family 27 member 2 |
| chr7_-_6221053 | 0.23 |
ENSDART00000173426
|
zgc:112234
|
zgc:112234 |
| chr23_-_1008468 | 0.22 |
ENSDART00000110588
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr3_-_1421391 | 0.20 |
ENSDART00000149709
|
smdt1a
|
single-pass membrane protein with aspartate-rich tail 1a |
| chr14_-_2873620 | 0.18 |
|
|
|
| chr10_+_4874800 | 0.18 |
ENSDART00000165942
|
palm2
|
paralemmin 2 |
| chr25_-_35820568 | 0.17 |
ENSDART00000152766
|
CR354435.4
|
Histone H2B 1/2 |
| chr25_+_25369539 | 0.17 |
ENSDART00000150537
|
rassf7a
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7a |
| chr4_-_44665935 | 0.15 |
ENSDART00000150524
|
si:ch211-162i8.4
|
si:ch211-162i8.4 |
| chr19_-_1376498 | 0.14 |
ENSDART00000158429
|
tmem42b
|
transmembrane protein 42b |
| chr10_+_34372283 | 0.10 |
ENSDART00000110121
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr3_+_15645543 | 0.10 |
|
|
|
| chr7_+_5846555 | 0.07 |
ENSDART00000083397
|
hist1h4l
|
histone 1, H4, like |
| chr6_+_30449742 | 0.00 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.8 | 2.3 | GO:0001502 | cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.2 | 2.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.2 | 0.7 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.2 | 0.6 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.1 | 0.6 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 | 0.6 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.4 | GO:0070861 | regulation of COPII vesicle coating(GO:0003400) regulation of protein exit from endoplasmic reticulum(GO:0070861) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.1 | 0.7 | GO:0006895 | phagolysosome assembly(GO:0001845) Golgi to endosome transport(GO:0006895) |
| 0.1 | 0.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.3 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 0.7 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 1.3 | GO:0043542 | endothelial cell migration(GO:0043542) |
| 0.0 | 1.6 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.9 | GO:0046890 | regulation of lipid biosynthetic process(GO:0046890) |
| 0.0 | 0.9 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.6 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 1.2 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.6 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 2.4 | GO:0000280 | nuclear division(GO:0000280) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.3 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 0.6 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.1 | 0.7 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 0.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.7 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 1.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 2.3 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 1.3 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.6 | GO:0016459 | myosin complex(GO:0016459) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 2.6 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.1 | 0.4 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.1 | 0.9 | GO:0005344 | oxygen transporter activity(GO:0005344) oxygen binding(GO:0019825) |
| 0.1 | 0.9 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 3.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.3 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.2 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 2.9 | GO:0051015 | actin filament binding(GO:0051015) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 2.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |