Project
DANIO-CODE
Navigation
Downloads
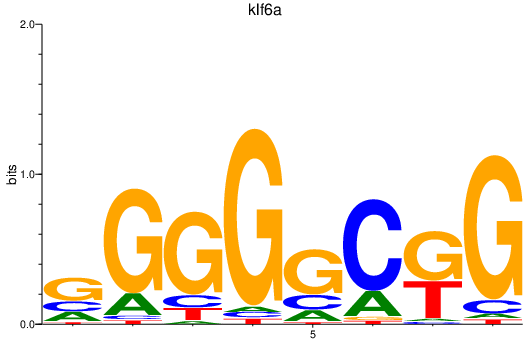
Results for klf6a
Z-value: 0.85
Transcription factors associated with klf6a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
klf6a
|
ENSDARG00000029072 | Kruppel-like factor 6a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| klf6a | dr10_dc_chr24_-_4117031_4117151 | -0.75 | 8.9e-04 | Click! |
Activity profile of klf6a motif
Sorted Z-values of klf6a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of klf6a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_30256477 | 4.93 |
ENSDART00000137193
|
slbp2
|
stem-loop binding protein 2 |
| chr9_-_55878791 | 2.50 |
|
|
|
| chr19_-_48327666 | 2.07 |
|
|
|
| KN149861v1_-_6661 | 1.61 |
|
|
|
| chr7_+_24855467 | 1.49 |
ENSDART00000077217
|
zgc:101765
|
zgc:101765 |
| chr22_-_111800 | 1.48 |
ENSDART00000163198
ENSDART00000168678 |
capzb
|
capping protein (actin filament) muscle Z-line, beta |
| chr7_+_45747622 | 1.47 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| KN149861v1_-_5655 | 1.47 |
|
|
|
| chr12_+_23939657 | 1.40 |
ENSDART00000021298
|
asb3
|
ankyrin repeat and SOCS box containing 3 |
| chr7_-_61373495 | 1.34 |
ENSDART00000098622
|
lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr23_-_43785497 | 1.26 |
ENSDART00000165963
|
ENSDARG00000102050
|
ENSDARG00000102050 |
| chr7_-_45579815 | 1.25 |
ENSDART00000170224
|
shcbp1
|
SHC SH2-domain binding protein 1 |
| chr6_-_6951460 | 1.21 |
ENSDART00000148879
|
nhej1
|
nonhomologous end-joining factor 1 |
| KN149861v1_-_6916 | 1.19 |
ENSDART00000169003
|
5_8S_rRNA
|
5.8S ribosomal RNA |
| chr6_-_12268874 | 1.15 |
|
|
|
| chr6_+_12268892 | 1.11 |
ENSDART00000171460
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr23_+_43785682 | 1.08 |
ENSDART00000172160
|
ENSDARG00000105093
|
ENSDARG00000105093 |
| chr20_+_27041190 | 1.08 |
ENSDART00000164027
|
cdca4
|
cell division cycle associated 4 |
| chr19_+_7717962 | 1.01 |
ENSDART00000112404
|
cgnb
|
cingulin b |
| KN149861v1_-_5543 | 0.99 |
|
|
|
| chr2_+_25212281 | 0.99 |
ENSDART00000078838
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr6_+_27523853 | 0.96 |
ENSDART00000128985
|
ryk
|
receptor-like tyrosine kinase |
| chr7_-_61373339 | 0.93 |
ENSDART00000148270
|
lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr22_-_26537845 | 0.93 |
|
|
|
| chr24_+_24581737 | 0.90 |
ENSDART00000137436
|
mtfr1
|
mitochondrial fission regulator 1 |
| chr12_+_23939869 | 0.87 |
ENSDART00000021298
|
asb3
|
ankyrin repeat and SOCS box containing 3 |
| chr20_+_27040570 | 0.86 |
ENSDART00000138369
|
cdca4
|
cell division cycle associated 4 |
| chr4_-_1809690 | 0.78 |
|
|
|
| chr21_-_2597788 | 0.73 |
|
|
|
| chr24_+_36564831 | 0.70 |
ENSDART00000168060
|
zgc:114120
|
zgc:114120 |
| chr13_-_11888896 | 0.70 |
ENSDART00000125430
|
pprc1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr17_+_6635602 | 0.70 |
ENSDART00000030773
|
foxo3a
|
forkhead box O3A |
| chr9_+_44921195 | 0.69 |
ENSDART00000145271
|
nckap1
|
NCK-associated protein 1 |
| chr16_+_36118041 | 0.68 |
ENSDART00000166040
|
sh3bp5b
|
SH3-domain binding protein 5b (BTK-associated) |
| chr13_-_11888944 | 0.67 |
ENSDART00000125430
|
pprc1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr17_-_20264870 | 0.67 |
|
|
|
| chr13_+_33578737 | 0.66 |
ENSDART00000161465
|
CABZ01087953.1
|
ENSDARG00000104106 |
| chr6_+_27524145 | 0.61 |
ENSDART00000079397
|
ryk
|
receptor-like tyrosine kinase |
| chr22_+_10202801 | 0.61 |
ENSDART00000105900
|
si:dkeyp-87e7.4
|
si:dkeyp-87e7.4 |
| chr17_-_20264762 | 0.60 |
|
|
|
| chr13_-_33527029 | 0.59 |
|
|
|
| chr6_-_60109646 | 0.56 |
ENSDART00000057463
ENSDART00000169188 |
pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr4_+_15007453 | 0.54 |
ENSDART00000146156
|
zc3hc1
|
zinc finger, C3HC-type containing 1 |
| chr6_+_54568649 | 0.51 |
ENSDART00000157142
|
tead3b
|
TEA domain family member 3 b |
| chr15_-_45522055 | 0.45 |
ENSDART00000113494
|
MB21D2
|
Mab-21 domain containing 2 |
| chr19_-_24971920 | 0.38 |
ENSDART00000132660
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr16_-_7875908 | 0.36 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr4_+_344355 | 0.34 |
ENSDART00000142225
ENSDART00000163436 |
tmem181
|
transmembrane protein 181 |
| chr7_-_23928945 | 0.31 |
|
|
|
| chr17_+_6635649 | 0.29 |
ENSDART00000030773
|
foxo3a
|
forkhead box O3A |
| chr13_-_11888864 | 0.29 |
ENSDART00000125430
|
pprc1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr6_+_54568388 | 0.28 |
ENSDART00000093199
|
tead3b
|
TEA domain family member 3 b |
| chr5_-_43219303 | 0.28 |
ENSDART00000022481
|
mccc2
|
methylcrotonoyl-CoA carboxylase 2 (beta) |
| chr21_+_43258389 | 0.28 |
ENSDART00000147026
|
shroom1
|
shroom family member 1 |
| chr14_-_24304628 | 0.24 |
|
|
|
| chr20_-_1248089 | 0.23 |
ENSDART00000027119
|
lats1
|
large tumor suppressor kinase 1 |
| chr16_+_34157948 | 0.23 |
ENSDART00000140552
|
tcea3
|
transcription elongation factor A (SII), 3 |
| chr9_-_3524962 | 0.23 |
ENSDART00000136599
|
cybrd1
|
cytochrome b reductase 1 |
| chr18_-_33832 | 0.22 |
ENSDART00000158957
ENSDART00000129125 |
pde8a
|
phosphodiesterase 8A |
| chr7_-_45747112 | 0.21 |
ENSDART00000172591
|
zgc:162297
|
zgc:162297 |
| chr15_+_44514665 | 0.19 |
|
|
|
| chr19_-_20522074 | 0.19 |
ENSDART00000043924
|
mpp6b
|
membrane protein, palmitoylated 6b (MAGUK p55 subfamily member 6) |
| chr22_+_26840335 | 0.19 |
ENSDART00000158756
|
crebbpa
|
CREB binding protein a |
| chr7_-_45747162 | 0.18 |
ENSDART00000172591
|
zgc:162297
|
zgc:162297 |
| chr14_-_32291093 | 0.12 |
ENSDART00000167282
ENSDART00000052938 |
atp11c
|
ATPase, Class VI, type 11C |
| chr3_-_30778523 | 0.07 |
ENSDART00000025046
|
ppp1caa
|
protein phosphatase 1, catalytic subunit, alpha isozyme a |
| chr7_-_61373398 | 0.05 |
ENSDART00000098622
|
lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr9_+_8475950 | 0.03 |
|
|
|
| chr20_+_54525614 | 0.03 |
ENSDART00000160409
|
arf6a
|
ADP-ribosylation factor 6a |
| chr6_-_18019342 | 0.02 |
ENSDART00000154941
|
ten1
|
TEN1 CST complex subunit |
| chr20_-_7186902 | 0.01 |
ENSDART00000012247
|
dhcr24
|
24-dehydrocholesterol reductase |
| chr3_-_15077819 | 0.00 |
ENSDART00000065807
|
kctd13
|
potassium channel tetramerization domain containing 13 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.9 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.2 | 1.0 | GO:0099558 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.2 | 1.9 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 1.0 | GO:1902743 | regulation of lamellipodium assembly(GO:0010591) regulation of lamellipodium organization(GO:1902743) |
| 0.1 | 1.4 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.1 | 0.6 | GO:0010991 | SMAD protein complex assembly(GO:0007183) regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.7 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.1 | 0.2 | GO:0070849 | response to epidermal growth factor(GO:0070849) cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.1 | 0.7 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 1.2 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.3 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 1.0 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.9 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 1.7 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 1.4 | GO:0000082 | G1/S transition of mitotic cell cycle(GO:0000082) |
| 0.0 | 1.0 | GO:0043523 | regulation of neuron apoptotic process(GO:0043523) |
| 0.0 | 1.3 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 0.6 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.2 | 1.5 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.2 | 1.0 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.3 | GO:1905202 | methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.1 | 0.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 2.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.7 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.6 | GO:0031901 | early endosome membrane(GO:0031901) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.9 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 0.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 1.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.3 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 0.2 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 1.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.7 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 1.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 1.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.6 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 2.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |