Project
DANIO-CODE
Navigation
Downloads
Results for klf7a+klf7b
Z-value: 0.46
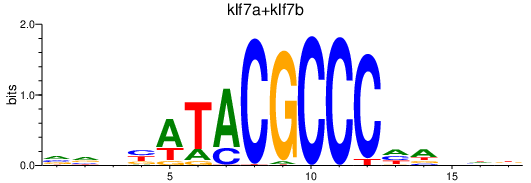
Transcription factors associated with klf7a+klf7b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
klf7b
|
ENSDARG00000043821 | Kruppel-like factor 7b |
|
klf7a
|
ENSDARG00000073857 | Kruppel-like factor 7a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| klf7b | dr10_dc_chr9_-_28588192_28588199 | 0.79 | 2.5e-04 | Click! |
| klf7a | dr10_dc_chr1_-_5796394_5796429 | 0.68 | 4.1e-03 | Click! |
Activity profile of klf7a+klf7b motif
Sorted Z-values of klf7a+klf7b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of klf7a+klf7b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_7531709 | 1.33 |
ENSDART00000104750
|
mllt11
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 11 |
| chr12_-_20228689 | 1.09 |
ENSDART00000066384
|
hbbe2
|
hemoglobin beta embryonic-2 |
| chr23_-_12079877 | 1.08 |
ENSDART00000147956
|
ENSDARG00000076364
|
ENSDARG00000076364 |
| chr2_-_23516930 | 1.06 |
ENSDART00000165355
|
prrx1a
|
paired related homeobox 1a |
| chr11_-_41357639 | 0.98 |
ENSDART00000055709
|
her2
|
hairy-related 2 |
| chr5_+_26195540 | 0.98 |
ENSDART00000145631
|
tcn2
|
transcobalamin II |
| chr25_+_35179025 | 0.95 |
ENSDART00000149768
|
kif21a
|
kinesin family member 21A |
| chr2_-_23517033 | 0.84 |
ENSDART00000041365
|
prrx1a
|
paired related homeobox 1a |
| chr19_+_31817286 | 0.82 |
ENSDART00000078459
|
tmem55a
|
transmembrane protein 55A |
| chr17_-_43725003 | 0.64 |
|
|
|
| chr1_+_13386937 | 0.56 |
ENSDART00000021693
|
ank2a
|
ankyrin 2a, neuronal |
| chr2_-_42185811 | 0.49 |
ENSDART00000140788
|
gbp1
|
guanylate binding protein 1 |
| chr2_-_42185736 | 0.48 |
ENSDART00000056460
|
gbp1
|
guanylate binding protein 1 |
| chr15_-_7327976 | 0.44 |
|
|
|
| chr20_+_18841440 | 0.41 |
ENSDART00000174532
|
fam167ab
|
family with sequence similarity 167, member Ab |
| chr20_+_13507202 | 0.38 |
ENSDART00000127350
|
enpp1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr24_+_21876490 | 0.36 |
ENSDART00000081234
|
ropn1l
|
rhophilin associated tail protein 1-like |
| chr19_-_7531649 | 0.35 |
ENSDART00000104750
|
mllt11
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 11 |
| chr2_-_23516991 | 0.32 |
ENSDART00000165355
|
prrx1a
|
paired related homeobox 1a |
| chr5_+_32191063 | 0.32 |
ENSDART00000077189
|
ier5l
|
immediate early response 5-like |
| chr9_+_748282 | 0.30 |
ENSDART00000004498
|
insig2
|
insulin induced gene 2 |
| chr1_-_43936717 | 0.29 |
ENSDART00000081835
|
slc43a1b
|
solute carrier family 43 (amino acid system L transporter), member 1b |
| chr1_-_10201135 | 0.26 |
ENSDART00000131744
|
dmd
|
dystrophin |
| chr21_+_8334453 | 0.22 |
ENSDART00000055336
|
dennd1a
|
DENN/MADD domain containing 1A |
| chr6_+_30681170 | 0.21 |
ENSDART00000112294
|
ttc22
|
tetratricopeptide repeat domain 22 |
| chr6_-_9798951 | 0.20 |
|
|
|
| chr2_+_3062119 | 0.19 |
ENSDART00000110416
|
eif2b5
|
eukaryotic translation initiation factor 2B, subunit 5 epsilon |
| chr4_-_28346021 | 0.19 |
ENSDART00000178149
|
celsr1a
|
cadherin EGF LAG seven-pass G-type receptor 1a |
| chr11_-_18073742 | 0.19 |
|
|
|
| chr1_-_43936611 | 0.18 |
ENSDART00000170912
|
slc43a1b
|
solute carrier family 43 (amino acid system L transporter), member 1b |
| chr2_-_23516868 | 0.18 |
ENSDART00000165355
|
prrx1a
|
paired related homeobox 1a |
| chr23_-_19534700 | 0.18 |
|
|
|
| chr7_-_37292692 | 0.17 |
ENSDART00000149052
|
nod2
|
nucleotide-binding oligomerization domain containing 2 |
| chr10_-_41235132 | 0.17 |
ENSDART00000134851
|
aak1b
|
AP2 associated kinase 1b |
| chr16_-_7911453 | 0.16 |
ENSDART00000108653
|
tcaim
|
T-cell activation inhibitor, mitochondrial |
| chr19_-_30834919 | 0.15 |
ENSDART00000043554
|
ppp1r10
|
protein phosphatase 1, regulatory subunit 10 |
| chr12_+_49071673 | 0.15 |
|
|
|
| chr11_-_2524674 | 0.14 |
ENSDART00000114079
|
nab2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr23_+_1176124 | 0.13 |
|
|
|
| chr15_-_7328008 | 0.11 |
|
|
|
| chr19_+_1743359 | 0.10 |
ENSDART00000166744
|
dennd3a
|
DENN/MADD domain containing 3a |
| chr17_-_37848208 | 0.08 |
ENSDART00000025853
|
zfp36l1a
|
zinc finger protein 36, C3H type-like 1a |
| chr11_-_18073851 | 0.07 |
|
|
|
| chr5_+_26195283 | 0.07 |
ENSDART00000053001
|
tcn2
|
transcobalamin II |
| chr20_+_20831085 | 0.04 |
ENSDART00000047662
|
ppp1r13bb
|
protein phosphatase 1, regulatory subunit 13Bb |
| chr25_+_21001071 | 0.04 |
ENSDART00000079012
|
rad52
|
RAD52 homolog, DNA repair protein |
| chr3_-_28741820 | 0.03 |
ENSDART00000138840
|
eef2kmt
|
eukaryotic elongation factor 2 lysine methyltransferase |
| chr18_-_26912259 | 0.02 |
ENSDART00000147735
|
si:dkey-24l11.2
|
si:dkey-24l11.2 |
| chr13_-_17333287 | 0.02 |
ENSDART00000145499
|
c13h10orf11
|
c13h10orf11 homolog (H. sapiens) |
| chr4_+_14728603 | 0.02 |
ENSDART00000131666
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr7_-_2230502 | 0.00 |
ENSDART00000173879
|
si:cabz01007794.1
|
si:cabz01007794.1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.4 | GO:0086019 | atrial cardiac muscle cell action potential(GO:0086014) cell-cell signaling involved in cardiac conduction(GO:0086019) atrial cardiac muscle cell to AV node cell signaling(GO:0086026) cell communication involved in cardiac conduction(GO:0086065) atrial cardiac muscle cell to AV node cell communication(GO:0086066) |
| 0.3 | 1.7 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) membrane depolarization(GO:0051899) regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.2 | 1.0 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.1 | 1.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.4 | GO:0055062 | regulation of fat cell differentiation(GO:0045598) phosphate ion homeostasis(GO:0055062) trivalent inorganic anion homeostasis(GO:0072506) |
| 0.1 | 0.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 0.2 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.1 | 1.0 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.3 | GO:0035437 | maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.5 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.2 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0005833 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 1.0 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.4 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.5 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |