Project
DANIO-CODE
Navigation
Downloads
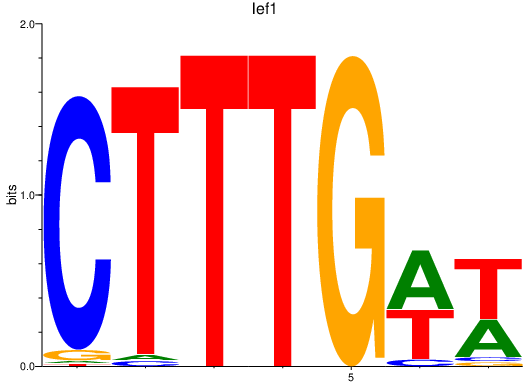
Results for lef1
Z-value: 2.96
Transcription factors associated with lef1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lef1
|
ENSDARG00000031894 | lymphoid enhancer-binding factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lef1 | dr10_dc_chr1_+_49170967_49170990 | -0.83 | 6.2e-05 | Click! |
Activity profile of lef1 motif
Sorted Z-values of lef1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of lef1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_16683931 | 12.02 |
ENSDART00000103262
ENSDART00000145068 ENSDART00000169619 ENSDART00000010526 |
fat1a
|
FAT atypical cadherin 1a |
| chr16_-_45950530 | 11.68 |
ENSDART00000060822
|
afp4
|
antifreeze protein type IV |
| chr14_-_7774262 | 8.07 |
ENSDART00000045109
|
zgc:92242
|
zgc:92242 |
| chr16_-_45943282 | 7.08 |
ENSDART00000133213
|
afp4
|
antifreeze protein type IV |
| chr4_-_11582061 | 6.53 |
ENSDART00000049066
|
net1
|
neuroepithelial cell transforming 1 |
| chr21_-_37509454 | 6.24 |
ENSDART00000175126
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr18_+_24935499 | 5.99 |
|
|
|
| chr9_-_3700395 | 5.75 |
ENSDART00000102900
|
sp5a
|
Sp5 transcription factor a |
| chr12_+_6432058 | 5.51 |
ENSDART00000066477
|
dkk1b
|
dickkopf WNT signaling pathway inhibitor 1b |
| chr1_-_50066633 | 5.26 |
|
|
|
| chr9_+_25520011 | 5.05 |
ENSDART00000167110
|
itm2bb
|
integral membrane protein 2Bb |
| chr2_-_8219329 | 5.01 |
ENSDART00000040209
|
ephb3a
|
eph receptor B3a |
| chr21_-_20291707 | 4.96 |
ENSDART00000079593
|
slc26a1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr15_-_24987099 | 4.93 |
|
|
|
| chr5_+_41680352 | 4.93 |
ENSDART00000142855
|
tbx6l
|
T-box 6, like |
| chr6_-_45903830 | 4.86 |
ENSDART00000025428
|
epha2a
|
eph receptor A2 a |
| chr24_-_37680649 | 4.74 |
ENSDART00000056286
|
h1f0
|
H1 histone family, member 0 |
| chr11_-_5868257 | 4.69 |
ENSDART00000104360
|
gamt
|
guanidinoacetate N-methyltransferase |
| chr16_+_23999029 | 4.61 |
ENSDART00000129525
|
apoa4b.1
|
apolipoprotein A-IV b, tandem duplicate 1 |
| chr19_+_5375413 | 4.51 |
ENSDART00000141237
|
si:dkeyp-113d7.10
|
si:dkeyp-113d7.10 |
| chr1_+_35933511 | 4.50 |
ENSDART00000010632
|
ednraa
|
endothelin receptor type Aa |
| chr5_+_34857986 | 4.28 |
ENSDART00000141239
|
erlin2
|
ER lipid raft associated 2 |
| chr7_+_44373815 | 4.27 |
ENSDART00000170721
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr25_-_22089794 | 4.25 |
ENSDART00000139110
|
pkp3a
|
plakophilin 3a |
| chr14_+_7626822 | 4.18 |
ENSDART00000109941
|
cxxc5b
|
CXXC finger protein 5b |
| chr10_+_32160464 | 4.16 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr21_-_28883441 | 4.13 |
ENSDART00000132884
|
cxxc5a
|
CXXC finger protein 5a |
| chr5_+_26925238 | 4.11 |
ENSDART00000051491
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr16_+_40610589 | 4.10 |
ENSDART00000038294
|
tp53inp1
|
tumor protein p53 inducible nuclear protein 1 |
| chr2_-_28446615 | 4.09 |
ENSDART00000179495
|
cdh6
|
cadherin 6 |
| chr5_+_49093250 | 3.99 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr23_+_19827551 | 3.98 |
ENSDART00000073442
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr17_+_52736192 | 3.97 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr6_-_32106882 | 3.92 |
ENSDART00000144772
|
BX664614.1
|
ENSDARG00000095311 |
| chr13_+_11305781 | 3.91 |
|
|
|
| chr6_-_43094573 | 3.90 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr22_-_22337268 | 3.90 |
|
|
|
| chr20_-_22576513 | 3.84 |
ENSDART00000103510
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr5_+_49093134 | 3.60 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr16_-_29779166 | 3.54 |
ENSDART00000067854
|
tnfaip8l2b
|
tumor necrosis factor, alpha-induced protein 8-like 2b |
| chr12_+_6007990 | 3.45 |
ENSDART00000091868
|
g6pca.2
|
glucose-6-phosphatase a, catalytic subunit, tandem duplicate 2 |
| chr1_+_9426040 | 3.43 |
ENSDART00000080576
|
lratb
|
lecithin retinol acyltransferase b (phosphatidylcholine-retinol O-acyltransferase) |
| chr23_+_32573474 | 3.43 |
ENSDART00000134811
|
si:dkey-261h17.1
|
si:dkey-261h17.1 |
| chr3_-_35928594 | 3.41 |
|
|
|
| chr25_+_18866796 | 3.40 |
ENSDART00000017299
|
tdg.1
|
thymine DNA glycosylase, tandem duplicate 1 |
| chr4_-_25075693 | 3.38 |
ENSDART00000025153
|
gata3
|
GATA binding protein 3 |
| chr18_-_36291570 | 3.38 |
|
|
|
| chr8_-_41194452 | 3.38 |
ENSDART00000165949
|
fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr3_-_29846530 | 3.35 |
ENSDART00000077111
|
hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr7_-_16776026 | 3.34 |
ENSDART00000022441
|
dbx1a
|
developing brain homeobox 1a |
| chr25_+_34135377 | 3.28 |
ENSDART00000157519
|
trpm1b
|
transient receptor potential cation channel, subfamily M, member 1b |
| chr12_-_9400344 | 3.27 |
ENSDART00000003932
|
erbb2
|
erb-b2 receptor tyrosine kinase 2 |
| chr15_+_23849554 | 3.25 |
ENSDART00000138375
|
ift20
|
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr9_-_14533551 | 3.25 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr11_-_29891067 | 3.23 |
ENSDART00000172106
|
scml2
|
sex comb on midleg-like 2 (Drosophila) |
| chr6_+_30441419 | 3.23 |
|
|
|
| chr4_+_57435 | 3.22 |
ENSDART00000169187
|
ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr8_-_16662185 | 3.21 |
ENSDART00000076542
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr24_-_4418454 | 3.21 |
ENSDART00000066835
|
fzd8a
|
frizzled class receptor 8a |
| chr7_-_54407681 | 3.20 |
ENSDART00000162795
|
ccnd1
|
cyclin D1 |
| chr1_-_50831155 | 3.19 |
ENSDART00000152719
|
spred2a
|
sprouty-related, EVH1 domain containing 2a |
| chr12_-_30988279 | 3.19 |
ENSDART00000122972
ENSDART00000005562 ENSDART00000031408 ENSDART00000125046 ENSDART00000009237 |
tcf7l2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr25_-_352985 | 3.14 |
ENSDART00000165705
|
szl
|
sizzled |
| chr6_-_40101104 | 3.13 |
ENSDART00000017402
|
ip6k2b
|
inositol hexakisphosphate kinase 2b |
| chr21_-_37286942 | 3.12 |
ENSDART00000100310
|
dbn1
|
drebrin 1 |
| chr11_+_7518953 | 3.10 |
ENSDART00000171813
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr4_-_25226680 | 3.07 |
ENSDART00000066932
ENSDART00000066933 ENSDART00000157714 |
itih2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr20_-_3302116 | 3.06 |
ENSDART00000123096
|
marcksa
|
myristoylated alanine-rich protein kinase C substrate a |
| chr1_-_7456961 | 3.04 |
ENSDART00000152295
|
FAM83G
|
family with sequence similarity 83 member G |
| chr4_+_18974767 | 3.04 |
ENSDART00000066973
|
impdh1b
|
IMP (inosine 5'-monophosphate) dehydrogenase 1b |
| chr6_-_13000193 | 3.04 |
|
|
|
| chr6_+_54703206 | 3.03 |
ENSDART00000074605
|
pkp1b
|
plakophilin 1b |
| chr2_+_48448974 | 3.03 |
ENSDART00000023040
|
hes6
|
hes family bHLH transcription factor 6 |
| chr9_-_11589126 | 3.02 |
ENSDART00000146832
|
cryba2b
|
crystallin, beta A2b |
| chr4_+_292818 | 3.02 |
ENSDART00000103659
|
msgn1
|
mesogenin 1 |
| chr6_+_6967174 | 3.01 |
ENSDART00000151311
|
ENSDARG00000058903
|
ENSDARG00000058903 |
| chr11_-_29316162 | 3.00 |
ENSDART00000163958
|
arhgef10la
|
Rho guanine nucleotide exchange factor (GEF) 10-like a |
| chr4_-_8610868 | 2.99 |
ENSDART00000067322
|
fbxl14b
|
F-box and leucine-rich repeat protein 14b |
| chr14_-_36523075 | 2.96 |
ENSDART00000109293
|
rnf130
|
ring finger protein 130 |
| chr16_-_32022177 | 2.95 |
ENSDART00000139664
|
styk1
|
serine/threonine/tyrosine kinase 1 |
| chr16_+_5625301 | 2.95 |
|
|
|
| chr20_+_6640497 | 2.92 |
ENSDART00000138361
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr22_+_16471319 | 2.89 |
ENSDART00000014330
|
ier5
|
immediate early response 5 |
| chr18_+_5899355 | 2.87 |
|
|
|
| chr14_-_40918015 | 2.87 |
ENSDART00000147389
|
tmem35
|
transmembrane protein 35 |
| chr13_-_31310439 | 2.87 |
ENSDART00000076571
|
rtn1a
|
reticulon 1a |
| chr6_-_23193752 | 2.85 |
ENSDART00000159749
|
usp43a
|
ubiquitin specific peptidase 43a |
| chr16_-_24697750 | 2.82 |
ENSDART00000163305
|
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr25_+_30746445 | 2.80 |
ENSDART00000156916
|
lsp1
|
lymphocyte-specific protein 1 |
| chr8_-_38357549 | 2.80 |
ENSDART00000129597
|
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr16_+_24697776 | 2.80 |
|
|
|
| chr24_-_23175007 | 2.79 |
ENSDART00000112256
|
zfhx4
|
zinc finger homeobox 4 |
| chr23_-_9924987 | 2.78 |
ENSDART00000005015
|
prkcbp1l
|
protein kinase C binding protein 1, like |
| chr8_-_51767046 | 2.78 |
ENSDART00000007090
|
tbx16
|
T-box 16 |
| chr18_+_45199548 | 2.77 |
|
|
|
| chr7_+_25466534 | 2.77 |
|
|
|
| chr25_-_12691849 | 2.74 |
ENSDART00000158551
|
slc7a5
|
solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
| chr15_-_15532980 | 2.74 |
ENSDART00000004220
ENSDART00000131259 |
rab34a
|
RAB34, member RAS oncogene family a |
| chr25_-_33670713 | 2.74 |
ENSDART00000125036
|
foxb1a
|
forkhead box B1a |
| chr11_-_17620732 | 2.73 |
ENSDART00000154627
|
eogt
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr21_-_25567978 | 2.73 |
ENSDART00000133134
|
efemp2b
|
EGF containing fibulin-like extracellular matrix protein 2b |
| chr16_-_39951493 | 2.72 |
|
|
|
| chr6_+_29800606 | 2.70 |
ENSDART00000017424
|
ptmaa
|
prothymosin, alpha a |
| chr12_-_5628401 | 2.69 |
ENSDART00000105884
|
itga3b
|
integrin, alpha 3b |
| chr13_+_27102308 | 2.67 |
ENSDART00000145901
|
rin2
|
Ras and Rab interactor 2 |
| chr24_-_33817169 | 2.66 |
ENSDART00000079292
|
cavin4b
|
caveolae associated protein 4b |
| chr23_+_6043862 | 2.64 |
|
|
|
| chr2_-_43110857 | 2.64 |
ENSDART00000098303
|
oc90
|
otoconin 90 |
| chr9_-_39190534 | 2.63 |
|
|
|
| chr23_-_35595271 | 2.62 |
ENSDART00000164616
|
tuba1c
|
tubulin, alpha 1c |
| chr14_-_25879853 | 2.62 |
ENSDART00000014149
|
fat2
|
FAT atypical cadherin 2 |
| chr4_+_12033088 | 2.61 |
ENSDART00000044154
|
tnnt2c
|
troponin T2c, cardiac |
| chr1_+_25662652 | 2.61 |
ENSDART00000113020
|
tet2
|
tet methylcytosine dioxygenase 2 |
| chr22_-_16015935 | 2.60 |
ENSDART00000062633
|
s1pr1
|
sphingosine-1-phosphate receptor 1 |
| chr5_+_48031920 | 2.60 |
ENSDART00000008043
ENSDART00000171438 |
adgrv1
|
adhesion G protein-coupled receptor V1 |
| KN149932v1_+_27584 | 2.60 |
|
|
|
| chr13_+_7575753 | 2.58 |
|
|
|
| chr15_+_28435937 | 2.57 |
ENSDART00000142298
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr3_+_22246697 | 2.56 |
ENSDART00000155597
|
arhgap27l
|
Rho GTPase activating protein 27, like |
| chr14_-_205130 | 2.56 |
ENSDART00000035581
ENSDART00000158725 ENSDART00000163549 |
OTOP1
|
otopetrin 1 |
| chr8_+_7740132 | 2.55 |
ENSDART00000171325
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr14_+_21999898 | 2.54 |
|
|
|
| chr7_-_54407461 | 2.50 |
ENSDART00000162795
|
ccnd1
|
cyclin D1 |
| chr9_-_42682501 | 2.47 |
ENSDART00000048320
|
tfpia
|
tissue factor pathway inhibitor a |
| chr23_+_23730717 | 2.46 |
|
|
|
| chr2_-_57244608 | 2.45 |
ENSDART00000149235
|
tcf3a
|
transcription factor 3a |
| chr13_-_39033893 | 2.44 |
ENSDART00000045434
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr11_-_36853345 | 2.44 |
ENSDART00000172074
|
NINJ1
|
ninjurin 1 |
| chr13_+_27102377 | 2.43 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr3_+_23591228 | 2.43 |
ENSDART00000012470
|
hoxb4a
|
homeobox B4a |
| chr13_+_11305846 | 2.40 |
|
|
|
| chr22_-_20101177 | 2.40 |
ENSDART00000138688
|
creb3l3a
|
cAMP responsive element binding protein 3-like 3a |
| chr1_+_27174549 | 2.39 |
ENSDART00000102337
|
dnajb14
|
DnaJ (Hsp40) homolog, subfamily B, member 14 |
| chr4_-_2187291 | 2.38 |
ENSDART00000150490
|
ENSDARG00000009262
|
ENSDARG00000009262 |
| chr23_+_27985224 | 2.37 |
ENSDART00000171859
|
ENSDARG00000100606
|
ENSDARG00000100606 |
| chr21_+_23916512 | 2.36 |
ENSDART00000145541
ENSDART00000065599 ENSDART00000112869 |
cadm1a
|
cell adhesion molecule 1a |
| chr18_-_23889256 | 2.35 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr15_-_11257445 | 2.35 |
|
|
|
| chr12_-_37560192 | 2.34 |
ENSDART00000140353
|
sdk2b
|
sidekick cell adhesion molecule 2b |
| chr4_-_73485028 | 2.34 |
ENSDART00000122898
|
phf21b
|
PHD finger protein 21B |
| chr10_+_18994733 | 2.33 |
ENSDART00000146517
|
dpysl2b
|
dihydropyrimidinase-like 2b |
| chr9_-_34068225 | 2.32 |
ENSDART00000088441
ENSDART00000150035 |
si:ch73-147f11.1
|
si:ch73-147f11.1 |
| chr21_-_41286846 | 2.32 |
ENSDART00000167339
|
msx2b
|
muscle segment homeobox 2b |
| chr21_-_39900518 | 2.31 |
ENSDART00000175055
|
CABZ01065291.1
|
ENSDARG00000107374 |
| chr24_+_4946542 | 2.30 |
ENSDART00000045813
|
zic4
|
zic family member 4 |
| chr7_-_38590391 | 2.29 |
ENSDART00000037361
ENSDART00000173629 ENSDART00000173953 |
phf21aa
|
PHD finger protein 21Aa |
| chr9_-_42894582 | 2.29 |
ENSDART00000144744
|
col5a2a
|
collagen, type V, alpha 2a |
| chr8_-_23591082 | 2.28 |
ENSDART00000025024
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr4_-_73484546 | 2.28 |
|
|
|
| chr21_+_45227200 | 2.27 |
ENSDART00000155681
ENSDART00000157136 |
tcf7
|
transcription factor 7 (T-cell specific, HMG-box) |
| chr5_+_42406222 | 2.26 |
ENSDART00000009182
|
aqp3a
|
aquaporin 3a |
| chr15_-_4154407 | 2.25 |
ENSDART00000090624
|
lpar6a
|
lysophosphatidic acid receptor 6a |
| chr7_+_48532749 | 2.25 |
ENSDART00000145375
|
cpt1aa
|
carnitine palmitoyltransferase 1Aa (liver) |
| chr11_-_42934175 | 2.24 |
ENSDART00000172929
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr9_+_25117850 | 2.24 |
|
|
|
| chr3_+_23587156 | 2.22 |
|
|
|
| chr15_-_26703761 | 2.22 |
ENSDART00000087632
|
ENSDARG00000079340
|
ENSDARG00000079340 |
| chr8_-_38168395 | 2.21 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr12_-_30662709 | 2.21 |
ENSDART00000126466
|
entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr8_+_14120313 | 2.18 |
ENSDART00000080832
|
si:dkey-6n6.2
|
si:dkey-6n6.2 |
| chr17_+_24613874 | 2.17 |
ENSDART00000093004
|
map4k3b
|
mitogen-activated protein kinase kinase kinase kinase 3b |
| chr1_-_35419335 | 2.17 |
ENSDART00000037516
|
znf827
|
zinc finger protein 827 |
| chr7_-_52142689 | 2.16 |
ENSDART00000110265
|
myzap
|
myocardial zonula adherens protein |
| chr10_+_7125001 | 2.16 |
ENSDART00000157987
|
psd3l
|
pleckstrin and Sec7 domain containing 3, like |
| chr25_-_18855316 | 2.16 |
ENSDART00000155927
|
si:ch211-68a17.7
|
si:ch211-68a17.7 |
| chr11_-_29586549 | 2.16 |
ENSDART00000079149
|
xk
|
X-linked Kx blood group (McLeod syndrome) |
| chr13_-_7434672 | 2.15 |
ENSDART00000159453
|
h2afy2
|
H2A histone family, member Y2 |
| chr18_-_23888988 | 2.14 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr11_+_30048913 | 2.13 |
|
|
|
| chr11_-_22966274 | 2.13 |
ENSDART00000003646
|
optc
|
opticin |
| chr21_+_10663517 | 2.13 |
ENSDART00000074833
|
rx3
|
retinal homeobox gene 3 |
| chr7_-_72500748 | 2.11 |
ENSDART00000160523
|
CU929444.1
|
ENSDARG00000099109 |
| chr24_+_18804086 | 2.11 |
ENSDART00000106186
|
prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr16_-_9785057 | 2.10 |
ENSDART00000113724
|
mal2
|
mal, T-cell differentiation protein 2 (gene/pseudogene) |
| chr18_+_40365208 | 2.09 |
ENSDART00000167134
|
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr6_-_43094926 | 2.09 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr17_-_12343975 | 2.09 |
ENSDART00000149551
|
emilin1b
|
elastin microfibril interfacer 1b |
| chr10_+_38582701 | 2.08 |
ENSDART00000144329
|
acer3
|
alkaline ceramidase 3 |
| chr17_-_20877606 | 2.07 |
ENSDART00000088106
|
ank3a
|
ankyrin 3a |
| chr15_-_31687 | 2.04 |
|
|
|
| chr25_-_5835981 | 2.03 |
ENSDART00000155751
|
nuak1b
|
NUAK family, SNF1-like kinase, 1b |
| chr9_-_23081250 | 2.03 |
|
|
|
| chr2_+_38178934 | 2.02 |
ENSDART00000044264
|
mmp14b
|
matrix metallopeptidase 14b (membrane-inserted) |
| chr16_+_33041823 | 2.01 |
ENSDART00000170157
|
prss35
|
protease, serine, 35 |
| chr25_+_19992389 | 2.01 |
ENSDART00000143441
|
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr17_+_24830545 | 2.01 |
ENSDART00000062917
|
cx35.4
|
connexin 35.4 |
| chr3_-_39287733 | 2.00 |
|
|
|
| chr1_-_18709814 | 2.00 |
ENSDART00000145224
|
rbm47
|
RNA binding motif protein 47 |
| chr21_-_26973987 | 1.99 |
ENSDART00000126542
|
ppp1r14ba
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Ba |
| chr20_-_37587804 | 1.98 |
ENSDART00000166106
|
si:ch211-202p1.5
|
si:ch211-202p1.5 |
| chr10_+_9716807 | 1.98 |
ENSDART00000064977
|
si:ch211-243g18.2
|
si:ch211-243g18.2 |
| chr8_+_33010788 | 1.97 |
ENSDART00000172672
|
angptl2b
|
angiopoietin-like 2b |
| chr16_+_5625552 | 1.97 |
|
|
|
| chr12_-_5628248 | 1.97 |
ENSDART00000105884
|
itga3b
|
integrin, alpha 3b |
| chr20_-_48677794 | 1.96 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr6_+_11594333 | 1.96 |
ENSDART00000109552
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr25_+_31547276 | 1.95 |
ENSDART00000090727
|
duox
|
dual oxidase |
| chr13_+_28601627 | 1.94 |
ENSDART00000015773
|
ldb1a
|
LIM domain binding 1a |
| chr18_-_23889025 | 1.94 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr6_+_54231519 | 1.94 |
ENSDART00000149542
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.6 | GO:0050810 | regulation of steroid biosynthetic process(GO:0050810) |
| 1.9 | 5.8 | GO:0048341 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 1.8 | 5.5 | GO:0061011 | hepatic duct development(GO:0061011) |
| 1.8 | 5.3 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 1.6 | 4.7 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 1.5 | 7.5 | GO:0060845 | lymphatic endothelial cell fate commitment(GO:0060838) venous endothelial cell differentiation(GO:0060843) venous endothelial cell fate commitment(GO:0060845) |
| 1.3 | 3.8 | GO:0048914 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 1.2 | 5.8 | GO:1903019 | negative regulation of glycoprotein biosynthetic process(GO:0010561) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) negative regulation of glycoprotein metabolic process(GO:1903019) |
| 1.1 | 4.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 1.1 | 3.3 | GO:2000583 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 1.0 | 15.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 1.0 | 5.1 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) mitotic G1 DNA damage checkpoint(GO:0031571) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 1.0 | 5.1 | GO:0045582 | positive regulation of T cell differentiation(GO:0045582) |
| 0.9 | 2.7 | GO:0061378 | associative learning(GO:0008306) visual learning(GO:0008542) mammillary axonal complex development(GO:0061373) mammillothalamic axonal tract development(GO:0061374) corpora quadrigemina development(GO:0061378) inferior colliculus development(GO:0061379) cell migration in diencephalon(GO:0061381) |
| 0.9 | 2.6 | GO:0051350 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclic nucleotide metabolic process(GO:0030800) negative regulation of cyclic nucleotide biosynthetic process(GO:0030803) negative regulation of nucleotide biosynthetic process(GO:0030809) negative regulation of cAMP metabolic process(GO:0030815) negative regulation of cAMP biosynthetic process(GO:0030818) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) negative regulation of purine nucleotide biosynthetic process(GO:1900372) |
| 0.9 | 6.0 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.8 | 5.0 | GO:0051963 | regulation of synapse assembly(GO:0051963) positive regulation of synapse assembly(GO:0051965) |
| 0.8 | 3.2 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.8 | 3.2 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.8 | 3.1 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.8 | 9.2 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.7 | 4.1 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.7 | 2.7 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.7 | 8.8 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.7 | 7.3 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.7 | 2.0 | GO:0014829 | phasic smooth muscle contraction(GO:0014821) vascular smooth muscle contraction(GO:0014829) endocrine process(GO:0050886) |
| 0.6 | 2.6 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.6 | 1.9 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.6 | 18.8 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.6 | 4.5 | GO:0042310 | vasoconstriction(GO:0042310) |
| 0.6 | 3.8 | GO:0003315 | heart rudiment formation(GO:0003315) |
| 0.6 | 3.8 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.6 | 1.7 | GO:1990359 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.6 | 1.1 | GO:0006699 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.6 | 3.3 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.5 | 2.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.5 | 2.6 | GO:0050482 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.5 | 2.6 | GO:0006211 | 5-methylcytosine catabolic process(GO:0006211) 5-methylcytosine metabolic process(GO:0019857) |
| 0.5 | 2.0 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.5 | 2.3 | GO:0033151 | V(D)J recombination(GO:0033151) |
| 0.5 | 4.1 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.4 | 1.8 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.4 | 2.5 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.4 | 2.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.4 | 3.2 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.4 | 1.2 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.4 | 1.2 | GO:1902024 | serine transport(GO:0032329) L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.4 | 2.0 | GO:2000742 | regulation of anterior head development(GO:2000742) |
| 0.4 | 1.2 | GO:1902893 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.4 | 1.1 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.4 | 1.1 | GO:0042423 | catechol-containing compound biosynthetic process(GO:0009713) catecholamine biosynthetic process(GO:0042423) |
| 0.4 | 2.2 | GO:0033986 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.4 | 3.9 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.3 | 2.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.3 | 1.3 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.3 | 2.0 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.3 | 1.9 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.3 | 3.5 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.3 | 1.6 | GO:0090317 | negative regulation of intracellular protein transport(GO:0090317) |
| 0.3 | 1.5 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.3 | 1.7 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.3 | 6.8 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.2 | 2.2 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.2 | 1.7 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.2 | 1.2 | GO:0090104 | pancreatic epsilon cell differentiation(GO:0090104) |
| 0.2 | 2.1 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.2 | 1.6 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.2 | 1.1 | GO:0006833 | water transport(GO:0006833) |
| 0.2 | 0.8 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.2 | 1.5 | GO:0021703 | rhombomere 5 development(GO:0021571) rhombomere 6 development(GO:0021572) locus ceruleus development(GO:0021703) |
| 0.2 | 0.6 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.2 | 0.6 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.2 | 3.7 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 4.5 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.2 | 0.8 | GO:0021547 | midbrain-hindbrain boundary initiation(GO:0021547) |
| 0.2 | 3.1 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.2 | 1.6 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.2 | 1.9 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.2 | 0.7 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.2 | 3.7 | GO:0071218 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.2 | 0.9 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.2 | 1.8 | GO:0061099 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.2 | 1.8 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.2 | 2.9 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 0.6 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 1.5 | GO:0072576 | liver morphogenesis(GO:0072576) |
| 0.1 | 1.0 | GO:1902534 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) late nucleophagy(GO:0044805) single-organism membrane invagination(GO:1902534) |
| 0.1 | 2.7 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.1 | 3.3 | GO:0060401 | cytosolic calcium ion transport(GO:0060401) calcium ion transport into cytosol(GO:0060402) |
| 0.1 | 1.0 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 2.3 | GO:0015807 | L-amino acid transport(GO:0015807) |
| 0.1 | 2.0 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 3.1 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.1 | 3.2 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.1 | 0.9 | GO:0090178 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 6.6 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.1 | 0.9 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 1.5 | GO:0001840 | neural plate development(GO:0001840) |
| 0.1 | 0.8 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 6.0 | GO:0048332 | mesoderm morphogenesis(GO:0048332) |
| 0.1 | 0.5 | GO:0055062 | regulation of fat cell differentiation(GO:0045598) phosphate ion homeostasis(GO:0055062) trivalent inorganic anion homeostasis(GO:0072506) |
| 0.1 | 0.7 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.6 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) ventricular cardiac muscle cell development(GO:0055015) |
| 0.1 | 0.7 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 2.2 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.9 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 1.6 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 1.1 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 0.6 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 0.5 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 1.5 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 3.2 | GO:0030835 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) actin filament capping(GO:0051693) |
| 0.1 | 1.2 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 2.5 | GO:0060119 | inner ear receptor cell development(GO:0060119) |
| 0.1 | 2.7 | GO:0007596 | blood coagulation(GO:0007596) hemostasis(GO:0007599) |
| 0.1 | 3.3 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.1 | 0.9 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 0.2 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 4.7 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.1 | 0.5 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.1 | 8.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 4.8 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 0.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 1.8 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 1.8 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.1 | 2.4 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 0.4 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.1 | 0.5 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 3.5 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 0.6 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.1 | 1.2 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.1 | 4.4 | GO:0010721 | negative regulation of cell development(GO:0010721) |
| 0.1 | 0.2 | GO:0060911 | cardiac cell fate commitment(GO:0060911) |
| 0.1 | 0.5 | GO:0021654 | rhombomere boundary formation(GO:0021654) |
| 0.1 | 5.3 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.1 | 2.3 | GO:0046148 | pigment biosynthetic process(GO:0046148) |
| 0.1 | 1.2 | GO:0039021 | pronephric glomerulus development(GO:0039021) |
| 0.1 | 1.9 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 0.1 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.1 | 0.6 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.1 | 3.0 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 0.2 | GO:0051026 | chiasma assembly(GO:0051026) meiotic metaphase plate congression(GO:0051311) |
| 0.1 | 0.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 1.5 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 5.6 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 2.3 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.7 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.7 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 | 1.3 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.4 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 1.3 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 1.0 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.3 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 1.1 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.5 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 0.0 | 0.3 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.3 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 4.8 | GO:0033674 | positive regulation of kinase activity(GO:0033674) |
| 0.0 | 0.2 | GO:0071938 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.0 | 0.2 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 4.5 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.2 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.4 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.4 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.0 | 0.7 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.2 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.5 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 1.0 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 1.1 | GO:0006874 | cellular calcium ion homeostasis(GO:0006874) |
| 0.0 | 0.3 | GO:0072599 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.0 | 0.8 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 3.2 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.0 | 1.9 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.1 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.4 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.8 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.1 | GO:0051029 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.0 | 0.4 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.2 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.8 | GO:1905037 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.9 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.2 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 0.5 | GO:0030641 | regulation of cellular pH(GO:0030641) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.2 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.8 | 2.3 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.6 | 1.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.5 | 2.6 | GO:0032420 | photoreceptor inner segment(GO:0001917) stereocilium(GO:0032420) |
| 0.4 | 2.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.4 | 8.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.4 | 7.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.3 | 3.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.3 | 1.3 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.3 | 1.2 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.2 | 4.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.2 | 1.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.2 | 2.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 2.9 | GO:0045178 | basal part of cell(GO:0045178) basal cortex(GO:0045180) |
| 0.2 | 8.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.2 | 4.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.2 | 3.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 4.8 | GO:0008305 | integrin complex(GO:0008305) |
| 0.2 | 1.8 | GO:0035060 | brahma complex(GO:0035060) |
| 0.2 | 1.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 2.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.4 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 3.9 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 0.9 | GO:0071256 | translocon complex(GO:0071256) |
| 0.1 | 0.5 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 1.1 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 20.6 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 0.7 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 4.1 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 1.0 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 2.9 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 0.8 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 1.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 2.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 0.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 0.5 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 5.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.9 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 2.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 4.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 0.3 | GO:0044304 | main axon(GO:0044304) |
| 0.1 | 0.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 2.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.2 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 1.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 1.6 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 6.5 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.7 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.8 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 6.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 22.4 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 4.2 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 1.0 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.7 | GO:0071004 | U2-type prespliceosome(GO:0071004) prespliceosome(GO:0071010) |
| 0.0 | 24.2 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.5 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.3 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 11.2 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 1.5 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.8 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 2.4 | GO:0099513 | supramolecular fiber(GO:0099512) polymeric cytoskeletal fiber(GO:0099513) |
| 0.0 | 0.1 | GO:0000812 | Swr1 complex(GO:0000812) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0039706 | co-receptor binding(GO:0039706) |
| 1.1 | 5.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 1.0 | 4.8 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.8 | 8.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.8 | 2.5 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.8 | 2.3 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) glycerol transmembrane transporter activity(GO:0015168) |
| 0.7 | 9.9 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.7 | 2.7 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.7 | 7.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.7 | 2.7 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.6 | 1.9 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.6 | 3.5 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.6 | 1.7 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.6 | 4.5 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.5 | 2.6 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.5 | 1.9 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.5 | 3.3 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.5 | 7.6 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.5 | 1.8 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.4 | 1.2 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.4 | 3.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.3 | 2.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.3 | 1.0 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.3 | 1.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.3 | 4.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.3 | 2.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.3 | 1.5 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.3 | 1.2 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.3 | 2.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.3 | 3.1 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.3 | 2.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.3 | 0.8 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.3 | 11.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.2 | 2.6 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.2 | 1.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.2 | 2.2 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.2 | 1.8 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.2 | 1.3 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.2 | 1.1 | GO:0015250 | water transmembrane transporter activity(GO:0005372) water channel activity(GO:0015250) |
| 0.2 | 2.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 7.7 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.2 | 1.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.2 | 7.1 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.2 | 0.7 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.2 | 2.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.7 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 2.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.9 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 0.9 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 4.1 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 2.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 0.4 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 1.2 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.9 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 0.6 | GO:0030546 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.1 | 4.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 2.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.4 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 1.0 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 6.8 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 4.5 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.1 | 3.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 2.2 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 4.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 2.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 4.8 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 1.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.1 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.5 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.1 | 1.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.4 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 0.4 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 1.8 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.1 | 3.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 2.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 30.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 1.9 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 1.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0034632 | retinol binding(GO:0019841) retinol transporter activity(GO:0034632) |
| 0.0 | 50.3 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 2.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 2.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) chemorepellent activity(GO:0045499) |
| 0.0 | 3.3 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 3.1 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 8.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 1.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.1 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 2.2 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 2.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.5 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 0.2 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 2.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0022841 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.9 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.4 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 1.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 8.2 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 3.0 | GO:0008757 | S-adenosylmethionine-dependent methyltransferase activity(GO:0008757) |
| 0.0 | 0.5 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.2 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.0 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.4 | 5.1 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.3 | 3.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 2.6 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.2 | 2.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 6.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.2 | 2.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 2.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 2.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 6.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 3.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 3.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 2.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 0.4 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 1.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 3.4 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 1.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 0.5 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 0.9 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 2.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.7 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.6 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.8 | 4.7 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.7 | 1.5 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.4 | 2.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.4 | 3.8 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.3 | 0.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 6.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 5.1 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.2 | 2.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 2.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 1.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.2 | 5.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 0.9 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 2.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.6 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.1 | 2.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 0.7 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.1 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.1 | 1.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 6.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 3.8 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 0.4 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 0.7 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 0.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.7 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 3.0 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 1.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 1.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 2.6 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.5 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.9 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.1 | REACTOME PI3K EVENTS IN ERBB2 SIGNALING | Genes involved in PI3K events in ERBB2 signaling |
| 0.0 | 0.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |