Project
DANIO-CODE
Navigation
Downloads
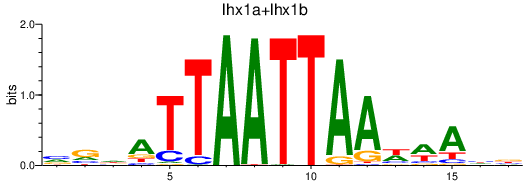
Results for lhx1a+lhx1b
Z-value: 0.31
Transcription factors associated with lhx1a+lhx1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lhx1b
|
ENSDARG00000007944 | LIM homeobox 1b |
|
lhx1a
|
ENSDARG00000014018 | LIM homeobox 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lhx1b | dr10_dc_chr5_+_55598181_55598220 | 0.77 | 4.7e-04 | Click! |
| lhx1a | dr10_dc_chr15_-_27778111_27778161 | 0.68 | 3.7e-03 | Click! |
Activity profile of lhx1a+lhx1b motif
Sorted Z-values of lhx1a+lhx1b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of lhx1a+lhx1b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_32686790 | 0.96 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr1_+_13779755 | 0.87 |
|
|
|
| chr20_-_45908435 | 0.67 |
ENSDART00000147637
|
fermt1
|
fermitin family member 1 |
| chr12_-_17590507 | 0.66 |
ENSDART00000010144
|
pvalb2
|
parvalbumin 2 |
| chr1_-_674449 | 0.66 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr21_-_1716495 | 0.58 |
ENSDART00000151049
|
onecut2
|
one cut homeobox 2 |
| chr19_-_5851328 | 0.51 |
ENSDART00000133106
|
si:ch211-264f5.6
|
si:ch211-264f5.6 |
| chr3_-_19218660 | 0.45 |
ENSDART00000132987
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr20_-_9107294 | 0.43 |
ENSDART00000140792
|
OMA1
|
OMA1 zinc metallopeptidase |
| chr17_+_15425559 | 0.42 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr4_+_9668755 | 0.40 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr24_+_35500964 | 0.37 |
ENSDART00000058571
|
snai2
|
snail family zinc finger 2 |
| chr4_+_16896821 | 0.37 |
ENSDART00000017726
|
etnk1
|
ethanolamine kinase 1 |
| chr9_-_14533551 | 0.34 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr17_+_49198580 | 0.31 |
ENSDART00000059255
ENSDART00000155599 |
zgc:113176
|
zgc:113176 |
| chr13_+_516287 | 0.30 |
ENSDART00000172469
ENSDART00000168523 ENSDART00000006892 |
tp53bp2a
|
tumor protein p53 binding protein, 2a |
| chr2_+_20748431 | 0.30 |
ENSDART00000137848
|
palmda
|
palmdelphin a |
| chr15_-_14616083 | 0.30 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr3_+_47735048 | 0.28 |
ENSDART00000083024
|
gpr146
|
G protein-coupled receptor 146 |
| chr2_+_38390887 | 0.26 |
ENSDART00000113111
|
psmb5
|
proteasome subunit beta 5 |
| chr24_+_21395671 | 0.26 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr8_+_21974863 | 0.26 |
|
|
|
| chr12_-_7902815 | 0.25 |
ENSDART00000088100
|
ank3b
|
ankyrin 3b |
| chr19_+_5562107 | 0.25 |
ENSDART00000082080
|
jupb
|
junction plakoglobin b |
| chr3_-_15849644 | 0.24 |
ENSDART00000160668
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr1_+_50395721 | 0.24 |
ENSDART00000134065
|
dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr5_-_31301280 | 0.23 |
ENSDART00000141446
|
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr10_-_11303185 | 0.23 |
ENSDART00000146727
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr9_-_8035360 | 0.22 |
|
|
|
| chr22_-_36561247 | 0.22 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr11_+_30416115 | 0.21 |
ENSDART00000161662
|
ttbk1a
|
tau tubulin kinase 1a |
| chr3_-_26657213 | 0.19 |
|
|
|
| chr11_-_41241693 | 0.19 |
ENSDART00000002556
|
mrto4
|
MRT4 homolog, ribosome maturation factor |
| chr2_-_49114158 | 0.19 |
|
|
|
| chr10_+_11303321 | 0.17 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr18_+_46384462 | 0.17 |
ENSDART00000024202
|
daw1
|
dynein assembly factor with WDR repeat domains 1 |
| chr2_-_2782967 | 0.17 |
ENSDART00000057458
|
CABZ01088149.1
|
ENSDARG00000039322 |
| chr16_-_28921444 | 0.17 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr7_+_25052687 | 0.17 |
ENSDART00000110347
|
cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr23_-_32043229 | 0.16 |
ENSDART00000134550
|
ormdl2
|
ORMDL sphingolipid biosynthesis regulator 2 |
| chr6_+_35069401 | 0.16 |
ENSDART00000082940
|
uhmk1
|
U2AF homology motif (UHM) kinase 1 |
| chr2_+_51087570 | 0.16 |
|
|
|
| chr20_-_28739099 | 0.16 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr14_-_33604914 | 0.16 |
ENSDART00000168546
|
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr9_+_34832049 | 0.15 |
ENSDART00000100735
ENSDART00000133996 |
shox
|
short stature homeobox |
| chr14_-_6901415 | 0.14 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr10_+_6120218 | 0.14 |
ENSDART00000166799
ENSDART00000157947 |
tln1
|
talin 1 |
| chr8_+_43334181 | 0.14 |
ENSDART00000038566
|
fam101a
|
family with sequence similarity 101, member A |
| chr11_-_25017693 | 0.13 |
|
|
|
| chr21_+_6046450 | 0.13 |
ENSDART00000131854
|
si:dkey-93m18.6
|
si:dkey-93m18.6 |
| chr23_-_4470717 | 0.12 |
ENSDART00000081823
|
si:ch73-142c19.1
|
si:ch73-142c19.1 |
| chr18_-_43890514 | 0.11 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr8_+_19471914 | 0.11 |
ENSDART00000159044
|
sec22bb
|
SEC22 homolog B, vesicle trafficking protein b |
| chr7_+_22539037 | 0.10 |
ENSDART00000020212
|
sf1
|
splicing factor 1 |
| chr13_+_516496 | 0.10 |
ENSDART00000172469
ENSDART00000168523 ENSDART00000006892 |
tp53bp2a
|
tumor protein p53 binding protein, 2a |
| chr17_+_25425967 | 0.10 |
ENSDART00000030691
|
clic4
|
chloride intracellular channel 4 |
| chr17_-_32471070 | 0.10 |
|
|
|
| chr13_+_516546 | 0.10 |
ENSDART00000172469
ENSDART00000168523 ENSDART00000006892 |
tp53bp2a
|
tumor protein p53 binding protein, 2a |
| chr13_-_29290894 | 0.09 |
ENSDART00000150228
|
chata
|
choline O-acetyltransferase a |
| chr5_+_3721680 | 0.09 |
ENSDART00000140537
|
dhrs11a
|
dehydrogenase/reductase (SDR family) member 11a |
| chr22_-_19527759 | 0.09 |
ENSDART00000105485
|
si:dkey-78l4.14
|
si:dkey-78l4.14 |
| chr5_+_18826505 | 0.09 |
|
|
|
| chr4_+_5246465 | 0.09 |
ENSDART00000137966
|
ccdc167
|
coiled-coil domain containing 167 |
| chr10_-_20545884 | 0.08 |
|
|
|
| chr3_+_3994248 | 0.08 |
|
|
|
| chr23_+_20714105 | 0.08 |
ENSDART00000054691
|
uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr22_-_12905648 | 0.08 |
ENSDART00000157820
|
mfsd6a
|
major facilitator superfamily domain containing 6a |
| chr3_+_50511676 | 0.08 |
ENSDART00000102202
|
ppap2d
|
phosphatidic acid phosphatase type 2D |
| chr22_-_12130441 | 0.08 |
ENSDART00000146785
|
tmem163b
|
transmembrane protein 163b |
| chr7_-_5366526 | 0.07 |
|
|
|
| chr3_+_27667194 | 0.07 |
ENSDART00000075100
|
carhsp1
|
calcium regulated heat stable protein 1 |
| chr5_+_45823481 | 0.07 |
|
|
|
| chr7_+_22538681 | 0.07 |
ENSDART00000020212
|
sf1
|
splicing factor 1 |
| chr2_+_1881022 | 0.07 |
ENSDART00000101038
|
tmie
|
transmembrane inner ear |
| chr14_-_1187074 | 0.06 |
ENSDART00000106672
|
arl9
|
ADP-ribosylation factor-like 9 |
| chr2_-_16548451 | 0.06 |
ENSDART00000152031
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr5_-_37494209 | 0.06 |
ENSDART00000084890
|
si:ch211-284e13.4
|
si:ch211-284e13.4 |
| chr6_-_50731538 | 0.06 |
ENSDART00000157153
|
pigu
|
phosphatidylinositol glycan anchor biosynthesis, class U |
| chr2_+_51087643 | 0.06 |
|
|
|
| chr2_+_25659945 | 0.06 |
ENSDART00000161386
|
fndc3ba
|
fibronectin type III domain containing 3Ba |
| chr12_+_1286911 | 0.05 |
ENSDART00000168225
ENSDART00000157467 |
pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr6_+_14854101 | 0.05 |
ENSDART00000087782
|
mrps9
|
mitochondrial ribosomal protein S9 |
| chr11_+_11284030 | 0.04 |
ENSDART00000026814
|
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr8_-_45752084 | 0.04 |
ENSDART00000025620
|
ppiaa
|
peptidylprolyl isomerase Aa (cyclophilin A) |
| chr15_-_21003820 | 0.04 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr16_+_54350976 | 0.03 |
ENSDART00000172622
ENSDART00000020033 |
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr13_-_51743395 | 0.03 |
|
|
|
| chr14_-_14302241 | 0.03 |
ENSDART00000162322
|
rab9b
|
RAB9B, member RAS oncogene family |
| chr8_-_12253026 | 0.03 |
ENSDART00000091612
|
dab2ipa
|
DAB2 interacting protein a |
| chr21_+_6858020 | 0.03 |
ENSDART00000139493
|
olfm1b
|
olfactomedin 1b |
| chr13_+_33138144 | 0.03 |
ENSDART00000002095
|
tmem39b
|
transmembrane protein 39B |
| chr15_-_36490729 | 0.03 |
|
|
|
| chr6_-_35068708 | 0.02 |
ENSDART00000170116
|
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr21_-_13875945 | 0.02 |
ENSDART00000143874
|
akna
|
AT-hook transcription factor |
| chr24_+_16402613 | 0.02 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr12_+_48497255 | 0.02 |
ENSDART00000168389
|
arhgap44
|
Rho GTPase activating protein 44 |
| chr22_-_4867538 | 0.02 |
ENSDART00000081801
|
ncl1
|
nicalin |
| chr1_+_9634016 | 0.02 |
ENSDART00000029774
|
tmem55bb
|
transmembrane protein 55Bb |
| chr24_+_26183653 | 0.01 |
ENSDART00000003884
|
mynn
|
myoneurin |
| chr7_-_52113622 | 0.01 |
ENSDART00000174186
|
wdr93
|
WD repeat domain 93 |
| KN150706v1_+_7627 | 0.01 |
|
|
|
| chr22_+_1691850 | 0.01 |
ENSDART00000170471
|
si:dkey-1b17.9
|
si:dkey-1b17.9 |
| chr7_+_24787720 | 0.01 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.1 | 0.2 | GO:0090153 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.5 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.2 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.3 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.2 | GO:0046822 | regulation of nucleocytoplasmic transport(GO:0046822) |
| 0.0 | 0.1 | GO:0019884 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.3 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.3 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.2 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.7 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.2 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.2 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.4 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |