Project
DANIO-CODE
Navigation
Downloads
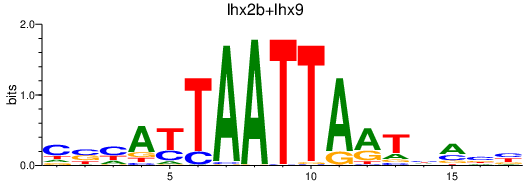
Results for lhx2b+lhx9
Z-value: 0.64
Transcription factors associated with lhx2b+lhx9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lhx2b
|
ENSDARG00000031222 | LIM homeobox 2b |
|
lhx9
|
ENSDARG00000056979 | LIM homeobox 9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lhx2b | dr10_dc_chr8_+_3026415_3026484 | 0.22 | 4.1e-01 | Click! |
| lhx9 | dr10_dc_chr22_-_23241074_23241101 | -0.12 | 6.7e-01 | Click! |
Activity profile of lhx2b+lhx9 motif
Sorted Z-values of lhx2b+lhx9 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of lhx2b+lhx9
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_26684617 | 1.26 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr16_-_45943282 | 1.14 |
ENSDART00000133213
|
afp4
|
antifreeze protein type IV |
| chr9_-_54126121 | 1.13 |
ENSDART00000126314
|
pcdh8
|
protocadherin 8 |
| chr24_-_38068071 | 1.11 |
ENSDART00000041805
|
metrn
|
meteorin, glial cell differentiation regulator |
| chr19_-_32175977 | 0.99 |
|
|
|
| chr19_-_27807312 | 0.94 |
|
|
|
| chr19_-_11239650 | 0.94 |
ENSDART00000146557
|
si:dkey-240h12.3
|
si:dkey-240h12.3 |
| chr8_+_28240085 | 0.93 |
ENSDART00000110857
|
fam212b
|
family with sequence similarity 212, member B |
| chr18_+_6551983 | 0.92 |
ENSDART00000160382
ENSDART00000171495 ENSDART00000160228 |
fam168a
|
family with sequence similarity 168, member A |
| chr13_-_22901327 | 0.89 |
ENSDART00000056523
|
hkdc1
|
hexokinase domain containing 1 |
| chr24_+_35500964 | 0.85 |
ENSDART00000058571
|
snai2
|
snail family zinc finger 2 |
| chr5_-_30114979 | 0.81 |
ENSDART00000016758
|
ftr82
|
finTRIM family, member 82 |
| chr16_-_9785057 | 0.78 |
ENSDART00000113724
|
mal2
|
mal, T-cell differentiation protein 2 (gene/pseudogene) |
| chr8_-_22945616 | 0.76 |
|
|
|
| chr2_-_38017902 | 0.73 |
ENSDART00000034790
|
pcp4l1
|
Purkinje cell protein 4 like 1 |
| chr20_+_109649 | 0.73 |
ENSDART00000022725
|
si:ch1073-155h21.1
|
si:ch1073-155h21.1 |
| chr7_-_17129072 | 0.72 |
|
|
|
| chr9_+_24891584 | 0.69 |
|
|
|
| chr21_+_25199691 | 0.67 |
ENSDART00000168140
ENSDART00000112783 |
tmem45b
|
transmembrane protein 45B |
| chr1_-_674449 | 0.66 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr11_-_25017693 | 0.66 |
|
|
|
| chr4_-_73485204 | 0.64 |
ENSDART00000122898
|
phf21b
|
PHD finger protein 21B |
| chr17_+_49198580 | 0.61 |
ENSDART00000059255
ENSDART00000155599 |
zgc:113176
|
zgc:113176 |
| chr3_-_38642067 | 0.61 |
ENSDART00000155518
|
znf281a
|
zinc finger protein 281a |
| chr16_-_11908084 | 0.61 |
ENSDART00000135408
|
cnfn
|
cornifelin |
| chr4_+_16896821 | 0.60 |
ENSDART00000017726
|
etnk1
|
ethanolamine kinase 1 |
| chr22_-_36561247 | 0.59 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr2_-_2782967 | 0.54 |
ENSDART00000057458
|
CABZ01088149.1
|
ENSDARG00000039322 |
| chr14_-_33604914 | 0.53 |
ENSDART00000168546
|
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr8_-_38308535 | 0.53 |
|
|
|
| chr17_-_51804002 | 0.52 |
ENSDART00000103350
ENSDART00000017329 |
numb
|
numb homolog (Drosophila) |
| chr20_+_19613133 | 0.51 |
ENSDART00000152548
ENSDART00000063696 |
atraid
|
all-trans retinoic acid-induced differentiation factor |
| chr14_-_6901415 | 0.51 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr25_+_35269590 | 0.51 |
ENSDART00000034737
|
cpne8
|
copine VIII |
| chr8_+_17132156 | 0.50 |
ENSDART00000134665
|
cenph
|
centromere protein H |
| chr13_+_7109810 | 0.50 |
ENSDART00000080975
|
tnfaip2b
|
tumor necrosis factor, alpha-induced protein 2b |
| chr9_+_21466934 | 0.49 |
ENSDART00000139620
|
lats2
|
large tumor suppressor kinase 2 |
| chr15_-_14616083 | 0.46 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr14_-_6901783 | 0.45 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr10_+_13042496 | 0.44 |
ENSDART00000158919
|
lpar1
|
lysophosphatidic acid receptor 1 |
| chr8_+_21974863 | 0.43 |
|
|
|
| chr16_-_12621624 | 0.43 |
ENSDART00000155829
|
cox6b2
|
cytochrome c oxidase subunit VIb polypeptide 2 |
| chr20_+_28465622 | 0.43 |
ENSDART00000146905
ENSDART00000103355 |
rhov
|
ras homolog family member V |
| chr10_-_43924675 | 0.43 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr5_+_45823481 | 0.43 |
|
|
|
| chr7_+_6517248 | 0.43 |
ENSDART00000102681
|
pnp5a
|
purine nucleoside phosphorylase 5a |
| chr9_-_8035360 | 0.43 |
|
|
|
| chr23_-_2957203 | 0.42 |
ENSDART00000165955
|
zhx3
|
zinc fingers and homeoboxes 3 |
| chr12_-_28248133 | 0.42 |
ENSDART00000016283
|
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr17_+_30352361 | 0.42 |
ENSDART00000076611
|
greb1
|
growth regulation by estrogen in breast cancer 1 |
| chr17_-_9794215 | 0.41 |
|
|
|
| chr16_+_29060022 | 0.41 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr18_+_17548213 | 0.39 |
ENSDART00000144960
|
nup93
|
nucleoporin 93 |
| chr2_-_10192027 | 0.38 |
ENSDART00000100709
|
imp3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr13_-_22901265 | 0.38 |
ENSDART00000056523
|
hkdc1
|
hexokinase domain containing 1 |
| chr20_-_9107294 | 0.38 |
ENSDART00000140792
|
OMA1
|
OMA1 zinc metallopeptidase |
| chr3_-_15849644 | 0.38 |
ENSDART00000160668
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr10_-_11303185 | 0.38 |
ENSDART00000146727
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr21_+_26684728 | 0.38 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr7_+_22539037 | 0.36 |
ENSDART00000020212
|
sf1
|
splicing factor 1 |
| chr16_-_42990753 | 0.36 |
ENSDART00000149317
|
hfe2
|
hemochromatosis type 2 |
| chr1_+_51706568 | 0.35 |
|
|
|
| chr8_+_7740132 | 0.34 |
ENSDART00000171325
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr22_-_12905648 | 0.34 |
ENSDART00000157820
|
mfsd6a
|
major facilitator superfamily domain containing 6a |
| chr5_+_27298136 | 0.34 |
ENSDART00000098604
|
adam28
|
ADAM metallopeptidase domain 28 |
| chr16_+_45192675 | 0.33 |
|
|
|
| chr23_-_33424398 | 0.32 |
ENSDART00000144831
ENSDART00000053416 |
si:ch211-226m16.2
|
si:ch211-226m16.2 |
| chr6_+_40924749 | 0.32 |
ENSDART00000133599
|
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr25_+_3091638 | 0.32 |
ENSDART00000104859
|
rccd1
|
RCC1 domain containing 1 |
| chr14_-_6831308 | 0.32 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr22_+_470536 | 0.32 |
ENSDART00000135403
|
nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr14_-_17257773 | 0.31 |
ENSDART00000082667
|
fgfrl1a
|
fibroblast growth factor receptor-like 1a |
| chr5_-_907020 | 0.31 |
|
|
|
| chr4_-_59767735 | 0.31 |
ENSDART00000159114
|
znf1129
|
zinc finger protein 1129 |
| chr23_-_21520413 | 0.31 |
ENSDART00000044080
ENSDART00000112929 |
her12
|
hairy-related 12 |
| chr13_-_51743395 | 0.31 |
|
|
|
| chr15_+_46813538 | 0.30 |
|
|
|
| chr19_+_43763483 | 0.30 |
ENSDART00000136695
|
yrk
|
Yes-related kinase |
| chr11_+_15935769 | 0.30 |
ENSDART00000158824
|
ctbs
|
chitobiase, di-N-acetyl- |
| chr17_-_11312185 | 0.29 |
ENSDART00000091159
|
adpgk2
|
ADP-dependent glucokinase 2 |
| chr14_-_45883839 | 0.29 |
|
|
|
| chr10_+_26570239 | 0.29 |
ENSDART00000139056
|
synj1
|
synaptojanin 1 |
| chr16_+_23516127 | 0.28 |
ENSDART00000004679
|
icn
|
ictacalcin |
| chr1_+_50395721 | 0.28 |
ENSDART00000134065
|
dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr23_-_42775849 | 0.28 |
ENSDART00000149944
|
gpx7
|
glutathione peroxidase 7 |
| chr25_+_10911620 | 0.28 |
ENSDART00000156589
ENSDART00000160922 |
mhc1lia
|
major histocompatibility complex class I LIA |
| chr2_-_49114158 | 0.28 |
|
|
|
| chr4_+_17666867 | 0.27 |
ENSDART00000066999
|
ccdc53
|
coiled-coil domain containing 53 |
| chr16_+_54350976 | 0.27 |
ENSDART00000172622
ENSDART00000020033 |
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr25_+_16078527 | 0.27 |
ENSDART00000176210
|
CR759887.4
|
ENSDARG00000108117 |
| chr8_+_25060320 | 0.27 |
ENSDART00000000744
|
sypl2b
|
synaptophysin-like 2b |
| chr4_-_73485366 | 0.26 |
ENSDART00000122898
|
phf21b
|
PHD finger protein 21B |
| chr7_+_58718614 | 0.26 |
ENSDART00000157873
|
hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr24_-_10928910 | 0.25 |
ENSDART00000127398
|
CR753886.1
|
ENSDARG00000090548 |
| chr8_-_38357549 | 0.25 |
ENSDART00000129597
|
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr23_-_30114528 | 0.25 |
ENSDART00000131209
|
ccdc187
|
coiled-coil domain containing 187 |
| chr7_+_21592861 | 0.24 |
ENSDART00000159626
|
si:dkey-85k7.7
|
si:dkey-85k7.7 |
| chr16_-_27743294 | 0.24 |
ENSDART00000145991
|
tbrg4
|
transforming growth factor beta regulator 4 |
| chr6_-_9471897 | 0.24 |
ENSDART00000039280
|
fzd7b
|
frizzled class receptor 7b |
| chr9_-_32803650 | 0.24 |
|
|
|
| chr10_+_29964045 | 0.24 |
ENSDART00000118838
|
SNORD14
|
Small nucleolar RNA SNORD14 |
| chr14_-_6831373 | 0.24 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr9_-_21677993 | 0.24 |
ENSDART00000121939
ENSDART00000080404 |
mphosph8
|
M-phase phosphoprotein 8 |
| chr5_-_6004819 | 0.23 |
ENSDART00000099570
|
tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr19_-_41933808 | 0.23 |
ENSDART00000062080
|
chrac1
|
chromatin accessibility complex 1 |
| chr16_-_31664756 | 0.23 |
ENSDART00000176939
|
phf20l1
|
PHD finger protein 20-like 1 |
| chr5_-_30115110 | 0.23 |
ENSDART00000016758
|
ftr82
|
finTRIM family, member 82 |
| chr3_+_27667194 | 0.23 |
ENSDART00000075100
|
carhsp1
|
calcium regulated heat stable protein 1 |
| chr13_-_22901481 | 0.23 |
ENSDART00000056523
|
hkdc1
|
hexokinase domain containing 1 |
| chr7_+_22538681 | 0.22 |
ENSDART00000020212
|
sf1
|
splicing factor 1 |
| chr17_+_42003279 | 0.22 |
ENSDART00000111537
|
kiz
|
kizuna centrosomal protein |
| chr21_+_17731439 | 0.22 |
ENSDART00000124173
|
rxraa
|
retinoid X receptor, alpha a |
| chr9_-_30744878 | 0.22 |
ENSDART00000079222
|
chaf1b
|
chromatin assembly factor 1, subunit B |
| chr19_+_7505348 | 0.22 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr6_+_19965515 | 0.21 |
ENSDART00000151240
|
si:dkey-156n14.3
|
si:dkey-156n14.3 |
| chr10_+_11303321 | 0.21 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr22_+_37696217 | 0.21 |
ENSDART00000007346
|
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr7_-_71290605 | 0.21 |
ENSDART00000111797
|
acer2
|
alkaline ceramidase 2 |
| chr21_-_26078089 | 0.21 |
ENSDART00000139320
|
nipal4
|
NIPA-like domain containing 4 |
| chr11_+_23522743 | 0.21 |
ENSDART00000121874
|
nfasca
|
neurofascin homolog (chicken) a |
| chr18_+_21419735 | 0.21 |
ENSDART00000144523
|
necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr2_+_1055716 | 0.21 |
ENSDART00000165477
|
AL935300.3
|
ENSDARG00000098661 |
| chr6_+_6640324 | 0.20 |
ENSDART00000150967
|
si:ch211-85n16.3
|
si:ch211-85n16.3 |
| chr16_-_22378805 | 0.20 |
ENSDART00000124718
|
aqp10a
|
aquaporin 10a |
| chr21_-_27584637 | 0.20 |
|
|
|
| chr5_-_30114910 | 0.20 |
ENSDART00000016758
|
ftr82
|
finTRIM family, member 82 |
| chr6_+_7376428 | 0.20 |
ENSDART00000057823
|
pa2g4a
|
proliferation-associated 2G4, a |
| chr13_+_28382422 | 0.19 |
ENSDART00000043117
|
fbxw4
|
F-box and WD repeat domain containing 4 |
| chr25_+_35771748 | 0.19 |
|
|
|
| chr8_+_36527535 | 0.19 |
ENSDART00000136418
ENSDART00000061378 |
sf3a1
|
splicing factor 3a, subunit 1 |
| chr9_-_32366320 | 0.19 |
ENSDART00000078568
|
sf3b1
|
splicing factor 3b, subunit 1 |
| chr2_+_20748431 | 0.19 |
ENSDART00000137848
|
palmda
|
palmdelphin a |
| chr7_-_50137683 | 0.18 |
ENSDART00000083346
|
hypk
|
huntingtin interacting protein K |
| chr7_-_5366526 | 0.18 |
|
|
|
| chr12_-_41128584 | 0.18 |
|
|
|
| chr12_-_18456073 | 0.18 |
ENSDART00000078166
|
zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr22_+_16509286 | 0.18 |
ENSDART00000083063
|
tal1
|
T-cell acute lymphocytic leukemia 1 |
| chr10_+_26691086 | 0.18 |
ENSDART00000079174
|
htatsf1
|
HIV-1 Tat specific factor 1 |
| chr3_-_30948297 | 0.18 |
ENSDART00000145636
|
ELOB (1 of many)
|
elongin B |
| chr23_+_11412329 | 0.17 |
ENSDART00000135406
|
chl1a
|
cell adhesion molecule L1-like a |
| chr18_-_5567053 | 0.17 |
ENSDART00000097960
|
myef2
|
myelin expression factor 2 |
| chr17_-_16414629 | 0.16 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| KN150544v1_+_53 | 0.16 |
|
|
|
| chr21_+_7737034 | 0.16 |
|
|
|
| chr8_-_19166630 | 0.16 |
|
|
|
| chr6_-_31374588 | 0.16 |
ENSDART00000027550
|
ak4
|
adenylate kinase 4 |
| chr20_-_52019761 | 0.16 |
ENSDART00000165076
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr19_+_44185325 | 0.16 |
ENSDART00000063870
|
rpl11
|
ribosomal protein L11 |
| chr25_-_20933145 | 0.16 |
ENSDART00000138985
ENSDART00000046298 |
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr3_+_3994248 | 0.16 |
|
|
|
| chr17_+_25425967 | 0.16 |
ENSDART00000030691
|
clic4
|
chloride intracellular channel 4 |
| chr20_+_25326042 | 0.15 |
ENSDART00000130494
|
moxd1
|
monooxygenase, DBH-like 1 |
| chr8_+_50499355 | 0.15 |
|
|
|
| chr4_-_935448 | 0.15 |
ENSDART00000171855
|
sim1b
|
single-minded family bHLH transcription factor 1b |
| chr3_-_57596004 | 0.15 |
ENSDART00000102062
|
timp2b
|
TIMP metallopeptidase inhibitor 2b |
| chr5_-_1131341 | 0.15 |
ENSDART00000168336
|
si:zfos-128g4.1
|
si:zfos-128g4.1 |
| chr16_+_24818799 | 0.14 |
ENSDART00000155217
|
si:dkey-79d12.4
|
si:dkey-79d12.4 |
| chr21_+_21158761 | 0.14 |
ENSDART00000058311
|
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr15_-_18226612 | 0.14 |
ENSDART00000012064
|
pih1d2
|
PIH1 domain containing 2 |
| chr10_+_29964164 | 0.14 |
ENSDART00000118838
|
SNORD14
|
Small nucleolar RNA SNORD14 |
| chr19_+_30226817 | 0.14 |
|
|
|
| chr1_+_51706536 | 0.14 |
|
|
|
| chr13_-_9543393 | 0.14 |
ENSDART00000041609
|
tm9sf3
|
transmembrane 9 superfamily member 3 |
| chr22_+_1335277 | 0.14 |
ENSDART00000169629
|
znf45l
|
zinc finger 45 like |
| chr9_-_33584043 | 0.14 |
ENSDART00000138844
|
caska
|
calcium/calmodulin-dependent serine protein kinase a |
| chr19_+_1106964 | 0.14 |
ENSDART00000165516
|
cfap20
|
cilia and flagella associated protein 20 |
| chr4_-_73485324 | 0.14 |
ENSDART00000122898
|
phf21b
|
PHD finger protein 21B |
| chr5_-_67395562 | 0.13 |
ENSDART00000165052
ENSDART00000018792 |
spag7
|
sperm associated antigen 7 |
| chr22_+_12306260 | 0.13 |
|
|
|
| chr9_-_30553051 | 0.13 |
ENSDART00000147030
|
sytl5
|
synaptotagmin-like 5 |
| chr24_-_14447136 | 0.13 |
|
|
|
| chr10_+_42693532 | 0.12 |
ENSDART00000010420
|
actr1
|
ARP1 actin related protein 1, centractin |
| chr17_-_30685367 | 0.12 |
ENSDART00000114358
|
zgc:194392
|
zgc:194392 |
| chr6_-_48419131 | 0.12 |
ENSDART00000090528
|
rhoca
|
ras homolog family member Ca |
| chr1_-_18059469 | 0.12 |
ENSDART00000140981
|
tbc1d1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr2_-_55673404 | 0.11 |
ENSDART00000158147
|
calr3b
|
calreticulin 3b |
| chr24_+_16402613 | 0.11 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr7_+_22538844 | 0.11 |
ENSDART00000020212
|
sf1
|
splicing factor 1 |
| chr17_+_19711153 | 0.11 |
|
|
|
| chr18_-_15451738 | 0.11 |
ENSDART00000091466
|
polr3b
|
polymerase (RNA) III (DNA directed) polypeptide B |
| chr23_+_4773894 | 0.11 |
ENSDART00000172739
|
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a |
| chr2_+_35745559 | 0.11 |
ENSDART00000002094
|
ankrd45
|
ankyrin repeat domain 45 |
| chr14_-_47129958 | 0.10 |
ENSDART00000172166
|
NDUFC1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr20_+_47639141 | 0.10 |
ENSDART00000043938
|
tram2
|
translocation associated membrane protein 2 |
| chr8_+_48953644 | 0.10 |
ENSDART00000132035
|
rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr9_+_7754307 | 0.10 |
ENSDART00000145853
|
ENSDARG00000006863
|
ENSDARG00000006863 |
| chr20_-_28453268 | 0.10 |
ENSDART00000128806
|
ino80
|
INO80 complex subunit |
| chr4_+_26064174 | 0.10 |
ENSDART00000171204
|
SCYL2
|
SCY1 like pseudokinase 2 |
| chr10_+_29963998 | 0.09 |
ENSDART00000118838
|
SNORD14
|
Small nucleolar RNA SNORD14 |
| chr19_-_7522951 | 0.09 |
ENSDART00000003544
|
gabpb2a
|
GA binding protein transcription factor, beta subunit 2a |
| chr9_-_33584087 | 0.09 |
ENSDART00000138844
|
caska
|
calcium/calmodulin-dependent serine protein kinase a |
| chr23_-_21520379 | 0.09 |
ENSDART00000044080
ENSDART00000112929 |
her12
|
hairy-related 12 |
| chr13_+_40311601 | 0.09 |
ENSDART00000148510
|
got1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr18_+_27769446 | 0.09 |
|
|
|
| chr15_-_43402935 | 0.09 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr23_+_20779487 | 0.09 |
ENSDART00000079538
|
ccdc30
|
coiled-coil domain containing 30 |
| chr8_+_21405788 | 0.09 |
ENSDART00000142758
|
si:dkey-163f12.10
|
si:dkey-163f12.10 |
| chr14_+_37317006 | 0.09 |
|
|
|
| chr6_+_14854101 | 0.09 |
ENSDART00000087782
|
mrps9
|
mitochondrial ribosomal protein S9 |
| chr25_-_20933295 | 0.08 |
ENSDART00000138985
ENSDART00000046298 |
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.2 | 1.6 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.1 | 0.8 | GO:0045056 | transcytosis(GO:0045056) |
| 0.1 | 0.5 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.1 | 1.5 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.1 | 0.6 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 0.3 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 0.4 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.4 | GO:0051292 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.4 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.5 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.1 | 0.3 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 0.3 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.2 | GO:2000815 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.1 | 0.4 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 1.2 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.1 | 0.2 | GO:0042423 | catechol-containing compound biosynthetic process(GO:0009713) catecholamine biosynthetic process(GO:0042423) |
| 0.0 | 0.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:2000253 | regulation of somitogenesis(GO:0014807) positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.5 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 1.0 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.0 | 0.2 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 1.1 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.3 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.0 | 0.2 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.0 | 0.9 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 0.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.7 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.4 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 0.3 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0042766 | nucleosome mobilization(GO:0042766) regulation of transcription from RNA polymerase II promoter in response to stress(GO:0043618) regulation of DNA-templated transcription in response to stress(GO:0043620) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.0 | 0.3 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.2 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.4 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.2 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.0 | 0.1 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.2 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 0.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.6 | GO:0005736 | DNA-directed RNA polymerase II, core complex(GO:0005665) DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.4 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.2 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.2 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.8 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.5 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0005536 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 0.7 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.2 | 0.6 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.2 | 0.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.5 | GO:0000215 | tRNA 2'-phosphotransferase activity(GO:0000215) 2'-phosphotransferase activity(GO:0008665) |
| 0.1 | 0.4 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.3 | GO:0102345 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.1 | 0.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.2 | GO:0015168 | polyol transmembrane transporter activity(GO:0015166) glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 1.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.0 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.5 | PID NOTCH PATHWAY | Notch signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.5 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.4 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.4 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.4 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.5 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |