Project
DANIO-CODE
Navigation
Downloads
Results for lhx5
Z-value: 0.66
Transcription factors associated with lhx5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lhx5
|
ENSDARG00000057936 | LIM homeobox 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lhx5 | dr10_dc_chr21_-_15833030_15833104 | -0.80 | 1.8e-04 | Click! |
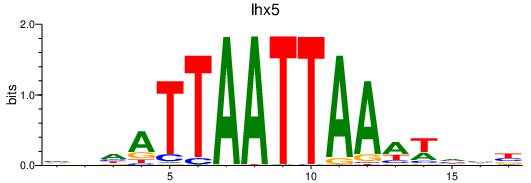
Activity profile of lhx5 motif
Sorted Z-values of lhx5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of lhx5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_44539778 | 2.38 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr19_-_1571878 | 2.05 |
|
|
|
| chr19_-_10411573 | 2.01 |
ENSDART00000171232
|
ccdc106b
|
coiled-coil domain containing 106b |
| chr21_+_22598805 | 1.93 |
ENSDART00000142495
|
BX510940.1
|
ENSDARG00000089088 |
| chr21_+_6289363 | 1.76 |
ENSDART00000138600
|
si:ch211-225g23.1
|
si:ch211-225g23.1 |
| chr11_+_17849608 | 1.58 |
ENSDART00000020283
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr16_+_14075878 | 1.47 |
ENSDART00000059926
|
zgc:162509
|
zgc:162509 |
| chr7_+_58910115 | 1.40 |
ENSDART00000172046
|
dok1b
|
docking protein 1b |
| chr7_-_73613531 | 1.39 |
ENSDART00000128137
|
zgc:92594
|
zgc:92594 |
| chr24_+_17115897 | 1.22 |
ENSDART00000129554
|
bmi1a
|
bmi1 polycomb ring finger oncogene 1a |
| chr2_-_17721810 | 1.21 |
ENSDART00000100201
ENSDART00000141188 |
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr7_-_26226924 | 1.15 |
ENSDART00000058910
|
sox19b
|
SRY (sex determining region Y)-box 19b |
| chr1_+_35253862 | 1.12 |
ENSDART00000139636
|
zgc:152968
|
zgc:152968 |
| chr23_+_28464194 | 1.11 |
ENSDART00000133736
|
CT025585.1
|
ENSDARG00000093306 |
| chr18_-_43890836 | 1.11 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr14_+_42109615 | 1.07 |
|
|
|
| chr18_+_35153749 | 1.07 |
ENSDART00000151073
|
si:ch211-195m9.3
|
si:ch211-195m9.3 |
| chr25_-_20933295 | 1.05 |
ENSDART00000138985
ENSDART00000046298 |
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr7_-_73613763 | 1.04 |
ENSDART00000128137
|
zgc:92594
|
zgc:92594 |
| chr5_-_3673698 | 1.03 |
|
|
|
| chr20_-_3220106 | 1.01 |
ENSDART00000123331
|
spint1b
|
serine peptidase inhibitor, Kunitz type 1 b |
| chr19_-_10411518 | 1.01 |
ENSDART00000171232
|
ccdc106b
|
coiled-coil domain containing 106b |
| chr6_-_50731449 | 0.99 |
ENSDART00000157153
|
pigu
|
phosphatidylinositol glycan anchor biosynthesis, class U |
| chr11_-_17668381 | 0.98 |
ENSDART00000080752
|
uba3
|
ubiquitin-like modifier activating enzyme 3 |
| chr14_-_16118942 | 0.96 |
|
|
|
| chr5_+_48013258 | 0.94 |
|
|
|
| chr6_+_40924559 | 0.88 |
ENSDART00000133599
|
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr20_-_3220392 | 0.85 |
ENSDART00000123331
|
spint1b
|
serine peptidase inhibitor, Kunitz type 1 b |
| chr5_-_48013023 | 0.85 |
ENSDART00000083229
|
mblac2
|
metallo-beta-lactamase domain containing 2 |
| chr23_+_28464298 | 0.84 |
ENSDART00000133736
|
CT025585.1
|
ENSDARG00000093306 |
| chr14_+_15845853 | 0.84 |
ENSDART00000160973
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr14_-_49777408 | 0.84 |
ENSDART00000166463
|
cnot6b
|
CCR4-NOT transcription complex, subunit 6b |
| chr3_+_40022244 | 0.82 |
ENSDART00000102526
|
gfer
|
growth factor, augmenter of liver regeneration (ERV1 homolog, S. cerevisiae) |
| chr5_-_48013093 | 0.81 |
ENSDART00000083229
|
mblac2
|
metallo-beta-lactamase domain containing 2 |
| chr8_-_53165501 | 0.81 |
ENSDART00000135982
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr16_-_32865452 | 0.79 |
|
|
|
| chr2_+_11094785 | 0.79 |
ENSDART00000091497
|
tceanc2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| chr22_+_2735606 | 0.78 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr25_-_20933145 | 0.78 |
ENSDART00000138985
ENSDART00000046298 |
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr23_+_28464642 | 0.78 |
ENSDART00000133736
|
CT025585.1
|
ENSDARG00000093306 |
| chr25_-_36616544 | 0.76 |
|
|
|
| chr21_-_44086464 | 0.75 |
ENSDART00000130833
|
FO704810.1
|
ENSDARG00000087492 |
| chr15_+_17162691 | 0.75 |
|
|
|
| chr15_-_5636737 | 0.74 |
ENSDART00000114410
|
wdr62
|
WD repeat domain 62 |
| chr7_-_26226628 | 0.74 |
ENSDART00000058910
|
sox19b
|
SRY (sex determining region Y)-box 19b |
| chr16_-_43061368 | 0.73 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr7_+_66660882 | 0.73 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr8_-_22720007 | 0.73 |
|
|
|
| chr5_-_31796734 | 0.73 |
ENSDART00000142095
|
fbxw2
|
F-box and WD repeat domain containing 2 |
| chr18_+_5066229 | 0.72 |
ENSDART00000157671
|
CABZ01080599.1
|
ENSDARG00000100626 |
| chr23_+_17583666 | 0.71 |
ENSDART00000054738
|
gid8b
|
GID complex subunit 8 homolog b (S. cerevisiae) |
| chr21_-_25765319 | 0.71 |
ENSDART00000101219
|
mettl27
|
methyltransferase like 27 |
| chr1_+_46808632 | 0.70 |
ENSDART00000027624
|
stn1
|
STN1, CST complex subunit |
| chr6_+_59580554 | 0.70 |
|
|
|
| chr8_+_36527653 | 0.70 |
ENSDART00000136418
ENSDART00000061378 |
sf3a1
|
splicing factor 3a, subunit 1 |
| chr10_-_43874597 | 0.69 |
ENSDART00000025366
|
cetn3
|
centrin 3 |
| chr15_-_44581550 | 0.68 |
|
|
|
| chr7_+_58910235 | 0.68 |
ENSDART00000172046
|
dok1b
|
docking protein 1b |
| chr14_+_37317006 | 0.68 |
|
|
|
| chr16_-_14075931 | 0.67 |
|
|
|
| chr9_+_24255311 | 0.64 |
ENSDART00000166303
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr4_-_5100307 | 0.63 |
ENSDART00000138817
|
tmem209
|
transmembrane protein 209 |
| chr23_+_29956616 | 0.63 |
ENSDART00000148706
|
aurkaip1
|
aurora kinase A interacting protein 1 |
| chr19_-_10411878 | 0.63 |
ENSDART00000136653
|
ccdc106b
|
coiled-coil domain containing 106b |
| chr23_+_17583383 | 0.62 |
ENSDART00000148457
|
gid8b
|
GID complex subunit 8 homolog b (S. cerevisiae) |
| chr20_+_25687135 | 0.62 |
ENSDART00000063122
|
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr8_+_39734109 | 0.62 |
ENSDART00000017153
|
hps4
|
Hermansky-Pudlak syndrome 4 |
| chr23_+_17583992 | 0.62 |
ENSDART00000054738
|
gid8b
|
GID complex subunit 8 homolog b (S. cerevisiae) |
| chr3_+_26212025 | 0.61 |
ENSDART00000163832
|
si:ch211-156b7.4
|
si:ch211-156b7.4 |
| chr7_-_73016194 | 0.61 |
|
|
|
| chr13_+_40347099 | 0.61 |
|
|
|
| chr22_-_28276824 | 0.60 |
ENSDART00000147686
|
si:dkey-222p3.1
|
si:dkey-222p3.1 |
| chr14_+_47229149 | 0.60 |
ENSDART00000136045
|
mgst2
|
microsomal glutathione S-transferase 2 |
| chr13_+_35402766 | 0.59 |
ENSDART00000163368
ENSDART00000075414 ENSDART00000112947 |
wdr27
|
WD repeat domain 27 |
| chr10_-_31129270 | 0.58 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr11_-_22210506 | 0.58 |
ENSDART00000146873
|
tmem183a
|
transmembrane protein 183A |
| chr10_-_31129217 | 0.58 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr19_+_5562075 | 0.57 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr8_+_50964745 | 0.57 |
ENSDART00000013870
|
ENSDARG00000007359
|
ENSDARG00000007359 |
| chr22_-_551324 | 0.57 |
|
|
|
| chr17_-_25813136 | 0.55 |
ENSDART00000148743
|
hhat
|
hedgehog acyltransferase |
| chr11_-_43821047 | 0.53 |
|
|
|
| chr4_-_5010148 | 0.53 |
ENSDART00000154025
|
strip2
|
striatin interacting protein 2 |
| chr24_-_14447655 | 0.53 |
|
|
|
| chr21_-_13026036 | 0.52 |
ENSDART00000135623
|
fam219aa
|
family with sequence similarity 219, member Aa |
| chr5_+_31475897 | 0.52 |
ENSDART00000144510
|
zmat5
|
zinc finger, matrin-type 5 |
| chr11_-_10472636 | 0.51 |
ENSDART00000081827
|
ect2
|
epithelial cell transforming 2 |
| chr25_-_13394261 | 0.51 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr1_-_50395003 | 0.50 |
ENSDART00000035150
|
spast
|
spastin |
| chr16_-_28723759 | 0.50 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr10_-_43874646 | 0.49 |
ENSDART00000025366
|
cetn3
|
centrin 3 |
| chr23_-_2957262 | 0.49 |
ENSDART00000165955
|
zhx3
|
zinc fingers and homeoboxes 3 |
| KN150040v1_-_6426 | 0.48 |
|
|
|
| chr15_+_34734212 | 0.48 |
ENSDART00000099776
|
tspan13a
|
tetraspanin 13a |
| chr19_+_1743359 | 0.46 |
ENSDART00000166744
|
dennd3a
|
DENN/MADD domain containing 3a |
| chr11_-_44539726 | 0.45 |
ENSDART00000173360
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr24_-_14447519 | 0.45 |
|
|
|
| chr11_-_10472565 | 0.45 |
ENSDART00000011087
|
ect2
|
epithelial cell transforming 2 |
| chr5_-_3673648 | 0.44 |
|
|
|
| chr4_-_75732882 | 0.44 |
|
|
|
| chr14_+_15845943 | 0.43 |
ENSDART00000159352
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr20_-_22532817 | 0.43 |
ENSDART00000177772
|
BX276097.1
|
ENSDARG00000108228 |
| chr7_-_57206977 | 0.42 |
ENSDART00000147036
|
sirt3
|
sirtuin 3 |
| chr18_+_8362265 | 0.42 |
ENSDART00000092053
|
chkb
|
choline kinase beta |
| chr3_-_48458042 | 0.42 |
ENSDART00000156822
|
cdip1
|
cell death-inducing p53 target 1 |
| chr5_-_57151021 | 0.41 |
ENSDART00000089961
|
sik2a
|
salt-inducible kinase 2a |
| chr6_+_40925259 | 0.41 |
ENSDART00000002728
ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr19_+_40482359 | 0.41 |
ENSDART00000151365
ENSDART00000140926 |
zmym4
|
zinc finger, MYM-type 4 |
| chr15_+_47633835 | 0.41 |
|
|
|
| chr11_+_5522377 | 0.41 |
ENSDART00000013203
|
cse1l
|
CSE1 chromosome segregation 1-like (yeast) |
| chr2_-_11094083 | 0.40 |
ENSDART00000100607
|
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr1_+_423654 | 0.39 |
ENSDART00000112730
|
tpp2
|
tripeptidyl peptidase 2 |
| chr17_-_15221715 | 0.39 |
ENSDART00000039165
|
styx
|
serine/threonine/tyrosine interacting protein |
| chr16_-_19636613 | 0.39 |
|
|
|
| chr5_+_13146987 | 0.39 |
ENSDART00000139199
|
h2afva
|
H2A histone family, member Va |
| chr5_+_20640396 | 0.38 |
ENSDART00000028087
|
cds2
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 |
| chr17_-_16414577 | 0.37 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr7_+_21006509 | 0.37 |
ENSDART00000052942
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr5_+_57151151 | 0.37 |
|
|
|
| chr12_-_43310712 | 0.37 |
ENSDART00000178532
ENSDART00000179029 |
BX321882.2
|
ENSDARG00000109101 |
| chr24_+_16402587 | 0.36 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr16_-_31386496 | 0.35 |
|
|
|
| chr3_+_18621758 | 0.35 |
ENSDART00000156747
|
AL772362.1
|
ENSDARG00000097552 |
| chr8_+_36467811 | 0.35 |
ENSDART00000098701
|
slc7a4
|
solute carrier family 7, member 4 |
| chr6_+_49552893 | 0.34 |
ENSDART00000022581
|
rab22a
|
RAB22A, member RAS oncogene family |
| chr20_+_22900302 | 0.34 |
ENSDART00000131132
|
scfd2
|
sec1 family domain containing 2 |
| chr9_-_43842377 | 0.34 |
ENSDART00000023684
|
cwc22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr15_-_43402935 | 0.34 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr23_+_17583294 | 0.34 |
ENSDART00000148457
|
gid8b
|
GID complex subunit 8 homolog b (S. cerevisiae) |
| chr18_-_23609418 | 0.33 |
ENSDART00000175930
|
CABZ01034364.1
|
ENSDARG00000107111 |
| chr23_-_2957169 | 0.33 |
ENSDART00000165955
|
zhx3
|
zinc fingers and homeoboxes 3 |
| chr9_-_35003703 | 0.32 |
|
|
|
| chr20_+_22900165 | 0.32 |
ENSDART00000058527
|
scfd2
|
sec1 family domain containing 2 |
| chr4_+_75469488 | 0.32 |
ENSDART00000144892
|
znf1009
|
zinc finger protein 1009 |
| chr17_+_22557630 | 0.32 |
ENSDART00000045099
|
yipf4
|
Yip1 domain family, member 4 |
| chr14_-_6901783 | 0.31 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr21_+_43177310 | 0.31 |
ENSDART00000121725
|
zcchc10
|
zinc finger, CCHC domain containing 10 |
| chr17_-_27030048 | 0.31 |
ENSDART00000050018
|
cnksr1
|
connector enhancer of kinase suppressor of Ras 1 |
| KN150040v1_+_6648 | 0.30 |
|
|
|
| chr19_+_7505348 | 0.30 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr11_-_28508152 | 0.29 |
|
|
|
| chr16_-_31664756 | 0.29 |
ENSDART00000176939
|
phf20l1
|
PHD finger protein 20-like 1 |
| chr6_-_40924459 | 0.29 |
ENSDART00000076097
|
sfi1
|
SFI1 centrin binding protein |
| chr13_+_35402382 | 0.28 |
ENSDART00000163368
|
wdr27
|
WD repeat domain 27 |
| chr18_-_605783 | 0.28 |
ENSDART00000157564
|
wtip
|
WT1 interacting protein |
| chr7_+_23752492 | 0.28 |
ENSDART00000141165
|
tinf2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr7_+_22538844 | 0.27 |
ENSDART00000020212
|
sf1
|
splicing factor 1 |
| chr10_+_1801745 | 0.26 |
|
|
|
| chr22_-_11585152 | 0.26 |
ENSDART00000063135
|
pcyt2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr17_-_41000670 | 0.26 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr20_-_5993068 | 0.24 |
ENSDART00000170468
|
cep128
|
centrosomal protein 128 |
| chr16_-_25914782 | 0.24 |
ENSDART00000086301
|
irge4
|
immunity-related GTPase family, e4 |
| chr7_+_28814408 | 0.24 |
ENSDART00000173960
|
acd
|
ACD, shelterin complex subunit and telomerase recruitment factor |
| chr11_+_17849528 | 0.23 |
ENSDART00000020283
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr6_+_59580339 | 0.23 |
|
|
|
| chr9_-_50304152 | 0.23 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr21_+_42722089 | 0.22 |
ENSDART00000177508
|
sh3pxd2b
|
SH3 and PX domains 2B |
| chr20_-_14116800 | 0.22 |
ENSDART00000152429
|
si:ch211-22i13.2
|
si:ch211-22i13.2 |
| chr6_+_14854074 | 0.22 |
ENSDART00000087782
|
mrps9
|
mitochondrial ribosomal protein S9 |
| chr11_+_35790782 | 0.22 |
ENSDART00000125221
|
CR933559.1
|
ENSDARG00000087939 |
| chr14_-_6831278 | 0.21 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr4_-_2370333 | 0.21 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr24_-_37178907 | 0.21 |
|
|
|
| chr21_-_17446442 | 0.20 |
|
|
|
| chr19_+_42486608 | 0.20 |
ENSDART00000151605
ENSDART00000166422 |
nfyc
|
nuclear transcription factor Y, gamma |
| chr9_-_50304120 | 0.20 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr24_-_32864493 | 0.20 |
|
|
|
| chr2_-_10192027 | 0.20 |
ENSDART00000100709
|
imp3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr8_+_36527535 | 0.20 |
ENSDART00000136418
ENSDART00000061378 |
sf3a1
|
splicing factor 3a, subunit 1 |
| chr23_+_4321502 | 0.19 |
ENSDART00000103747
|
srsf6a
|
serine/arginine-rich splicing factor 6a |
| chr17_-_37447869 | 0.18 |
ENSDART00000148160
|
crip1
|
cysteine-rich protein 1 |
| chr9_+_31204464 | 0.18 |
|
|
|
| chr17_-_16414629 | 0.17 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr7_+_21592861 | 0.17 |
ENSDART00000159626
|
si:dkey-85k7.7
|
si:dkey-85k7.7 |
| chr2_+_42223071 | 0.17 |
ENSDART00000134203
|
vcpip1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr14_-_6831373 | 0.17 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr22_-_36561217 | 0.16 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr16_+_51597530 | 0.16 |
|
|
|
| chr18_+_15964146 | 0.15 |
|
|
|
| chr25_-_27279272 | 0.15 |
ENSDART00000148121
|
zgc:153935
|
zgc:153935 |
| chr19_+_46567459 | 0.15 |
ENSDART00000164055
|
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr7_+_23752651 | 0.14 |
ENSDART00000141165
|
tinf2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr1_+_51087450 | 0.14 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr5_+_20640468 | 0.14 |
ENSDART00000028087
|
cds2
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 |
| chr16_-_35462083 | 0.12 |
ENSDART00000170048
|
taf12
|
TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr1_+_35253821 | 0.12 |
ENSDART00000179634
|
zgc:152968
|
zgc:152968 |
| chr17_-_32471070 | 0.12 |
|
|
|
| chr16_-_28723722 | 0.12 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr3_-_32686757 | 0.10 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr20_+_51391607 | 0.09 |
|
|
|
| chr14_-_32291093 | 0.09 |
ENSDART00000167282
ENSDART00000052938 |
atp11c
|
ATPase, Class VI, type 11C |
| chr1_+_9633924 | 0.08 |
ENSDART00000029774
|
tmem55bb
|
transmembrane protein 55Bb |
| chr6_-_9471897 | 0.08 |
ENSDART00000039280
|
fzd7b
|
frizzled class receptor 7b |
| chr18_+_8362131 | 0.08 |
ENSDART00000092053
|
chkb
|
choline kinase beta |
| chr16_+_51597409 | 0.08 |
|
|
|
| chr4_+_2551928 | 0.08 |
ENSDART00000103371
ENSDART00000171405 |
zdhhc17
|
zinc finger, DHHC-type containing 17 |
| chr7_+_58427742 | 0.07 |
ENSDART00000073640
|
plag1
|
pleiomorphic adenoma gene 1 |
| chr21_-_13875945 | 0.06 |
ENSDART00000143874
|
akna
|
AT-hook transcription factor |
| chr3_-_11657881 | 0.06 |
ENSDART00000127157
|
hlfa
|
hepatic leukemia factor a |
| chr25_+_35771748 | 0.06 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.3 | 1.2 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.2 | 1.8 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.2 | 0.6 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.2 | 2.8 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.4 | GO:0043393 | regulation of protein binding(GO:0043393) |
| 0.1 | 0.5 | GO:0043243 | positive regulation of protein complex disassembly(GO:0043243) |
| 0.1 | 1.2 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.1 | 0.6 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.1 | 0.5 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 1.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 0.7 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 1.4 | GO:0016233 | telomere capping(GO:0016233) |
| 0.1 | 1.0 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 2.3 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.1 | 0.2 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.1 | 0.4 | GO:0007589 | body fluid secretion(GO:0007589) |
| 0.1 | 0.4 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 1.0 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.5 | GO:1902547 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.0 | 0.6 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 1.9 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 0.2 | GO:0048308 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.0 | 0.5 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.0 | 0.2 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.8 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.1 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.6 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.1 | GO:0045761 | regulation of cAMP metabolic process(GO:0030814) regulation of cAMP biosynthetic process(GO:0030817) regulation of adenylate cyclase activity(GO:0045761) |
| 0.0 | 0.4 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.7 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:0019430 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 0.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.6 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.6 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.6 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1990879 | CST complex(GO:1990879) |
| 0.2 | 0.6 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 1.2 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 1.0 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.7 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 2.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.7 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 0.8 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.3 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.1 | 0.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 1.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.8 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.9 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 1.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.0 | 0.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.3 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.5 | GO:0030496 | midbody(GO:0030496) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.2 | 0.7 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.2 | 0.6 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.2 | 0.5 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.1 | 0.8 | GO:0016972 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.1 | 0.4 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.1 | 1.8 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 1.2 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.1 | 0.6 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 1.8 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.7 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.1 | 0.5 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.1 | 0.5 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 0.7 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 2.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 2.8 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.7 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 1.2 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.4 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.1 | GO:0000215 | tRNA 2'-phosphotransferase activity(GO:0000215) 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 1.2 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.1 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.0 | 0.6 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 1.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 2.5 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.7 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 0.7 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.7 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.8 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.7 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.0 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.8 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.5 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 1.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |