Project
DANIO-CODE
Navigation
Downloads
Results for lhx6
Z-value: 1.03
Transcription factors associated with lhx6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lhx6
|
ENSDARG00000006896 | LIM homeobox 6 |
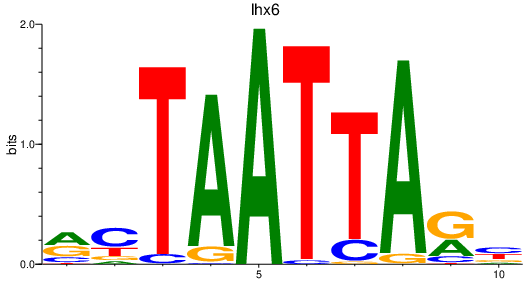
Activity profile of lhx6 motif
Sorted Z-values of lhx6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of lhx6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_23759076 | 7.52 |
ENSDART00000145894
|
zgc:195245
|
zgc:195245 |
| KN150456v1_-_19515 | 5.15 |
ENSDART00000168786
|
h1m
|
linker histone H1M |
| chr11_-_44539778 | 4.34 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr10_-_34058331 | 4.21 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr10_-_21404605 | 3.81 |
ENSDART00000125167
|
avd
|
avidin |
| chr24_-_14446593 | 3.69 |
|
|
|
| chr10_+_33449922 | 3.67 |
ENSDART00000115379
ENSDART00000163458 ENSDART00000078012 |
zgc:153345
|
zgc:153345 |
| chr20_-_14218080 | 3.60 |
ENSDART00000104032
|
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr12_-_14104939 | 3.54 |
ENSDART00000152742
|
buc2l
|
bucky ball 2-like |
| chr23_+_2786407 | 3.42 |
ENSDART00000066086
|
zgc:114123
|
zgc:114123 |
| chr5_+_37303599 | 3.32 |
ENSDART00000097754
ENSDART00000162470 |
tmprss4b
|
transmembrane protease, serine 4b |
| chr19_-_18664720 | 3.22 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| chr5_+_36168475 | 2.90 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr24_+_19270877 | 2.85 |
|
|
|
| chr21_-_32027717 | 2.80 |
ENSDART00000131651
|
ENSDARG00000073961
|
ENSDARG00000073961 |
| chr20_-_6542402 | 2.72 |
ENSDART00000054653
|
mcm3l
|
MCM3 minichromosome maintenance deficient 3 (S. cerevisiae), like |
| chr2_-_26941084 | 2.67 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr25_+_5845303 | 2.60 |
ENSDART00000163948
|
ENSDARG00000053246
|
ENSDARG00000053246 |
| chr4_+_4825461 | 2.48 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr20_+_14218237 | 2.41 |
ENSDART00000044937
|
kcns3b
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3b |
| chr3_+_17783319 | 2.24 |
ENSDART00000104299
|
cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr12_-_33256671 | 2.23 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr5_-_68011971 | 2.20 |
ENSDART00000141699
|
mepce
|
methylphosphate capping enzyme |
| chr11_+_23799984 | 2.19 |
|
|
|
| chr1_-_22617455 | 2.13 |
ENSDART00000137567
|
smim14
|
small integral membrane protein 14 |
| chr21_-_32747592 | 2.11 |
|
|
|
| chr2_+_6341404 | 2.03 |
ENSDART00000076700
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr14_-_8634381 | 2.00 |
ENSDART00000129030
|
zgc:153681
|
zgc:153681 |
| chr12_-_33256754 | 1.96 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr2_-_26941232 | 1.93 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr3_-_29725539 | 1.92 |
|
|
|
| chr2_-_15656155 | 1.90 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| KN150663v1_-_3381 | 1.89 |
|
|
|
| chr1_-_18118467 | 1.87 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr19_+_39689450 | 1.85 |
|
|
|
| chr18_-_43890836 | 1.85 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr23_-_10850891 | 1.80 |
|
|
|
| chr19_+_10742387 | 1.76 |
ENSDART00000091813
|
ago3b
|
argonaute RISC catalytic component 3b |
| chr5_+_19429620 | 1.71 |
ENSDART00000088819
|
ankrd13a
|
ankyrin repeat domain 13A |
| chr6_-_40715613 | 1.68 |
ENSDART00000153702
|
si:ch211-157b11.12
|
si:ch211-157b11.12 |
| chr21_+_19040595 | 1.68 |
ENSDART00000145969
|
BX000991.1
|
ENSDARG00000092282 |
| chr12_-_33257026 | 1.67 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr12_+_33257120 | 1.64 |
|
|
|
| chr1_+_13244109 | 1.62 |
ENSDART00000157563
|
noctb
|
nocturnin b |
| chr20_-_29961498 | 1.58 |
ENSDART00000132278
|
rnf144ab
|
ring finger protein 144ab |
| chr23_+_28464194 | 1.56 |
ENSDART00000133736
|
CT025585.1
|
ENSDARG00000093306 |
| chr7_-_33414221 | 1.56 |
|
|
|
| chr22_-_21873054 | 1.56 |
|
|
|
| chr12_-_33256934 | 1.52 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr1_+_50547385 | 1.47 |
ENSDART00000132141
|
btbd3a
|
BTB (POZ) domain containing 3a |
| chr3_-_20944579 | 1.46 |
ENSDART00000153739
|
nlk1
|
nemo-like kinase, type 1 |
| chr20_-_29961589 | 1.45 |
ENSDART00000132278
|
rnf144ab
|
ring finger protein 144ab |
| chr5_+_19429500 | 1.45 |
ENSDART00000168868
|
ankrd13a
|
ankyrin repeat domain 13A |
| chr10_-_32550351 | 1.45 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| KN149710v1_+_38638 | 1.43 |
|
|
|
| chr25_-_13394261 | 1.42 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr2_-_26940965 | 1.41 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr19_-_18664670 | 1.39 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| chr16_+_47283253 | 1.38 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr14_-_6901209 | 1.38 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr16_+_42567707 | 1.36 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr12_-_33256599 | 1.34 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr6_+_29763568 | 1.33 |
ENSDART00000151784
|
ptmaa
|
prothymosin, alpha a |
| chr20_-_14218236 | 1.32 |
ENSDART00000168434
|
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr10_-_2944190 | 1.32 |
ENSDART00000132526
|
marveld2a
|
MARVEL domain containing 2a |
| chr5_-_18502344 | 1.31 |
ENSDART00000137022
ENSDART00000090494 ENSDART00000165701 |
golga3
|
golgin A3 |
| chr1_+_35253862 | 1.31 |
ENSDART00000139636
|
zgc:152968
|
zgc:152968 |
| chr8_-_22720007 | 1.31 |
|
|
|
| chr5_+_6391432 | 1.28 |
ENSDART00000170564
ENSDART00000086666 |
stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr14_-_15777250 | 1.26 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr10_-_35313462 | 1.24 |
ENSDART00000139107
|
prr11
|
proline rich 11 |
| chr15_+_21327206 | 1.22 |
ENSDART00000101000
|
gkup
|
glucuronokinase with putative uridyl pyrophosphorylase |
| chr10_+_33450122 | 1.22 |
ENSDART00000115379
ENSDART00000163458 ENSDART00000078012 |
zgc:153345
|
zgc:153345 |
| chr5_+_6005049 | 1.20 |
|
|
|
| chr9_-_35824470 | 1.20 |
ENSDART00000140356
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr24_+_39630741 | 1.15 |
ENSDART00000132939
|
dcun1d3
|
defective in cullin neddylation 1 domain containing 3 |
| chr19_+_10742594 | 1.12 |
ENSDART00000091813
|
ago3b
|
argonaute RISC catalytic component 3b |
| chr23_-_33692244 | 1.11 |
|
|
|
| chr10_+_35313772 | 1.09 |
ENSDART00000028940
|
styxl1
|
serine/threonine/tyrosine interacting-like 1 |
| chr20_+_29306863 | 1.07 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr20_+_29306677 | 1.07 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr6_+_59580554 | 1.05 |
|
|
|
| chr14_+_23420053 | 1.05 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr16_+_47283374 | 1.03 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr20_-_16271738 | 1.02 |
ENSDART00000012476
|
coa7
|
cytochrome c oxidase assembly factor 7 |
| chr6_-_19556029 | 1.02 |
ENSDART00000136019
|
ppp1r12c
|
protein phosphatase 1, regulatory subunit 12C |
| chr23_+_28464143 | 1.01 |
ENSDART00000133736
|
CT025585.1
|
ENSDARG00000093306 |
| KN150040v1_-_6426 | 0.99 |
|
|
|
| chr1_+_18118735 | 0.99 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr20_-_23327126 | 0.97 |
ENSDART00000153308
|
dcun1d4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) |
| chr19_+_31998646 | 0.96 |
ENSDART00000132182
|
gmnn
|
geminin, DNA replication inhibitor |
| KN150030v1_-_22613 | 0.95 |
ENSDART00000175410
|
CABZ01079802.1
|
ENSDARG00000106760 |
| chr10_+_2948868 | 0.93 |
ENSDART00000147918
|
zfyve16
|
zinc finger, FYVE domain containing 16 |
| chr20_-_37910887 | 0.93 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr11_-_44539726 | 0.91 |
ENSDART00000173360
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr18_+_8362265 | 0.91 |
ENSDART00000092053
|
chkb
|
choline kinase beta |
| chr21_+_43333462 | 0.88 |
ENSDART00000109620
|
sept8a
|
septin 8a |
| chr6_-_40715579 | 0.88 |
ENSDART00000157113
ENSDART00000154810 |
si:ch211-157b11.12
|
si:ch211-157b11.12 |
| chr19_+_10742344 | 0.88 |
ENSDART00000091813
|
ago3b
|
argonaute RISC catalytic component 3b |
| chr13_-_24695520 | 0.87 |
ENSDART00000142745
|
slka
|
STE20-like kinase a |
| chr21_-_20291586 | 0.86 |
ENSDART00000079593
|
slc26a1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr15_+_28477741 | 0.85 |
ENSDART00000057257
|
pitpnaa
|
phosphatidylinositol transfer protein, alpha a |
| chr8_-_21110262 | 0.84 |
ENSDART00000143192
|
cpt2
|
carnitine palmitoyltransferase 2 |
| chr6_-_8009055 | 0.80 |
ENSDART00000151358
|
rgl3a
|
ral guanine nucleotide dissociation stimulator-like 3a |
| chr1_+_50547341 | 0.76 |
ENSDART00000132141
|
btbd3a
|
BTB (POZ) domain containing 3a |
| chr21_-_39521698 | 0.76 |
ENSDART00000020174
|
dynll2b
|
dynein, light chain, LC8-type 2b |
| chr2_+_10209233 | 0.75 |
ENSDART00000160304
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr20_-_23327219 | 0.75 |
ENSDART00000142721
|
dcun1d4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) |
| chr23_-_2957262 | 0.74 |
ENSDART00000165955
|
zhx3
|
zinc fingers and homeoboxes 3 |
| chr20_+_6542597 | 0.72 |
|
|
|
| chr24_-_17261929 | 0.72 |
ENSDART00000153858
|
BX005392.2
|
ENSDARG00000096996 |
| chr23_+_11781792 | 0.72 |
|
|
|
| chr5_+_37303707 | 0.71 |
ENSDART00000097754
ENSDART00000162470 |
tmprss4b
|
transmembrane protease, serine 4b |
| chr4_+_4825628 | 0.71 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr20_+_35176069 | 0.71 |
|
|
|
| chr25_-_28630138 | 0.70 |
|
|
|
| chr21_-_14079056 | 0.70 |
ENSDART00000111659
|
dfnb31a
|
deafness, autosomal recessive 31a |
| chr24_+_16402587 | 0.69 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr24_-_14447825 | 0.69 |
|
|
|
| KN150040v1_+_6648 | 0.68 |
|
|
|
| chr6_-_3821922 | 0.68 |
ENSDART00000171944
|
slc25a12
|
solute carrier family 25 (aspartate/glutamate carrier), member 12 |
| chr24_-_14447655 | 0.68 |
|
|
|
| chr4_-_1952230 | 0.68 |
ENSDART00000135749
|
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr14_+_23419864 | 0.68 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr5_+_21564036 | 0.67 |
ENSDART00000045574
|
shisa2a
|
shisa family member 2a |
| chr14_+_29601073 | 0.67 |
ENSDART00000143763
|
fam149a
|
family with sequence similarity 149 member A |
| chr8_-_21110183 | 0.66 |
ENSDART00000143192
|
cpt2
|
carnitine palmitoyltransferase 2 |
| chr12_+_22459177 | 0.66 |
ENSDART00000171725
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr21_-_32747544 | 0.65 |
|
|
|
| chr22_-_551324 | 0.65 |
|
|
|
| chr5_-_18502442 | 0.65 |
ENSDART00000165639
|
golga3
|
golgin A3 |
| chr22_-_21872864 | 0.65 |
ENSDART00000158501
|
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr5_+_25133592 | 0.65 |
ENSDART00000098467
|
abhd17b
|
abhydrolase domain containing 17B |
| chr11_-_27378184 | 0.63 |
ENSDART00000157337
|
CR931782.1
|
ENSDARG00000097455 |
| chr2_-_38380883 | 0.63 |
ENSDART00000088026
|
prmt5
|
protein arginine methyltransferase 5 |
| chr4_+_13587809 | 0.62 |
ENSDART00000138201
|
tnpo3
|
transportin 3 |
| chr12_+_22459218 | 0.62 |
ENSDART00000171725
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr6_-_19556121 | 0.61 |
ENSDART00000136019
|
ppp1r12c
|
protein phosphatase 1, regulatory subunit 12C |
| chr2_+_6341345 | 0.61 |
ENSDART00000058256
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr21_-_14079001 | 0.61 |
ENSDART00000111659
|
dfnb31a
|
deafness, autosomal recessive 31a |
| chr17_+_8642373 | 0.60 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr13_+_36797201 | 0.59 |
ENSDART00000026313
|
tmx1
|
thioredoxin-related transmembrane protein 1 |
| chr23_-_10851353 | 0.59 |
ENSDART00000055038
|
rybpa
|
RING1 and YY1 binding protein a |
| chr6_-_19556328 | 0.59 |
ENSDART00000074256
|
ppp1r12c
|
protein phosphatase 1, regulatory subunit 12C |
| chr15_-_43402935 | 0.59 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr14_+_8634323 | 0.58 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr16_-_42105636 | 0.58 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr19_-_19806070 | 0.58 |
ENSDART00000165206
|
smarcc1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1b |
| chr14_-_6901783 | 0.58 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr8_-_25015215 | 0.57 |
ENSDART00000170511
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr25_-_12236333 | 0.56 |
ENSDART00000174863
ENSDART00000179042 |
BX323452.1
|
ENSDARG00000107774 |
| chr16_+_42567668 | 0.56 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr20_+_29306945 | 0.55 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr13_-_42274444 | 0.55 |
ENSDART00000043069
|
march5
|
membrane-associated ring finger (C3HC4) 5 |
| chr23_-_2957169 | 0.55 |
ENSDART00000165955
|
zhx3
|
zinc fingers and homeoboxes 3 |
| chr24_-_14447519 | 0.54 |
|
|
|
| chr7_+_8492338 | 0.51 |
|
|
|
| chr7_-_68229115 | 0.51 |
|
|
|
| chr21_+_18238176 | 0.49 |
ENSDART00000144322
|
wdr5
|
WD repeat domain 5 |
| chr24_-_14446522 | 0.49 |
|
|
|
| chr7_+_17656192 | 0.49 |
ENSDART00000077113
|
mta2
|
metastasis associated 1 family, member 2 |
| chr9_-_50304152 | 0.48 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr16_-_26982451 | 0.47 |
ENSDART00000078119
|
ino80c
|
INO80 complex subunit C |
| chr23_+_40240121 | 0.47 |
|
|
|
| chr16_-_42105733 | 0.46 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr14_+_29601252 | 0.46 |
ENSDART00000143763
|
fam149a
|
family with sequence similarity 149 member A |
| chr9_-_746424 | 0.46 |
ENSDART00000082300
|
usp37
|
ubiquitin specific peptidase 37 |
| chr9_-_50304120 | 0.44 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr12_+_26785950 | 0.42 |
ENSDART00000087329
|
znf438
|
zinc finger protein 438 |
| chr7_+_17656099 | 0.42 |
ENSDART00000077113
|
mta2
|
metastasis associated 1 family, member 2 |
| chr17_-_8413036 | 0.42 |
ENSDART00000154713
|
fzd3b
|
frizzled class receptor 3b |
| chr7_+_72925910 | 0.40 |
ENSDART00000175267
ENSDART00000179638 |
CABZ01067170.1
|
ENSDARG00000106453 |
| chr7_-_26260802 | 0.40 |
ENSDART00000121698
|
senp3b
|
SUMO1/sentrin/SMT3 specific peptidase 3b |
| chr20_-_48772949 | 0.40 |
ENSDART00000170894
|
mgst3a
|
microsomal glutathione S-transferase 3a |
| chr7_+_72030256 | 0.38 |
ENSDART00000172021
|
tollip
|
toll interacting protein |
| chr11_+_43924766 | 0.38 |
ENSDART00000179206
|
gnb4b
|
guanine nucleotide binding protein (G protein), beta polypeptide 4b |
| chr24_-_24859135 | 0.38 |
ENSDART00000136860
|
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr8_-_23759198 | 0.38 |
ENSDART00000145894
|
zgc:195245
|
zgc:195245 |
| chr17_+_8054784 | 0.37 |
ENSDART00000133301
|
slc18b1
|
solute carrier family 18, subfamily B, member 1 |
| chr5_-_19429016 | 0.36 |
ENSDART00000170344
|
git2a
|
G protein-coupled receptor kinase interacting ArfGAP 2a |
| chr23_+_37807201 | 0.36 |
ENSDART00000129531
|
CR381544.1
|
ENSDARG00000088187 |
| chr6_+_59580339 | 0.35 |
|
|
|
| chr1_-_39384698 | 0.34 |
ENSDART00000147317
|
cntf
|
ciliary neurotrophic factor |
| chr11_-_29520809 | 0.33 |
ENSDART00000079117
|
plekha3
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3 |
| chr14_-_15776937 | 0.33 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr23_+_37807239 | 0.33 |
ENSDART00000129531
|
CR381544.1
|
ENSDARG00000088187 |
| chr16_+_24046996 | 0.30 |
|
|
|
| chr13_-_48884956 | 0.29 |
|
|
|
| chr7_+_28814408 | 0.29 |
ENSDART00000173960
|
acd
|
ACD, shelterin complex subunit and telomerase recruitment factor |
| chr18_+_8362131 | 0.29 |
ENSDART00000092053
|
chkb
|
choline kinase beta |
| chr13_+_22346304 | 0.29 |
ENSDART00000137220
|
ldb3a
|
LIM domain binding 3a |
| chr15_+_28477893 | 0.29 |
ENSDART00000057257
|
pitpnaa
|
phosphatidylinositol transfer protein, alpha a |
| chr17_-_36904594 | 0.29 |
|
|
|
| chr7_-_38540389 | 0.28 |
|
|
|
| chr5_-_25133456 | 0.28 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr3_+_53792981 | 0.28 |
ENSDART00000178486
|
olfm2a
|
olfactomedin 2a |
| chr17_+_8642428 | 0.27 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr17_-_41000670 | 0.25 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr11_+_11284030 | 0.24 |
ENSDART00000026814
|
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr19_+_21783204 | 0.23 |
ENSDART00000024639
|
tshz1
|
teashirt zinc finger homeobox 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.6 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.9 | 3.4 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.4 | 2.2 | GO:1900182 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.4 | 2.7 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.4 | 1.1 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.4 | 5.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 3.8 | GO:0072091 | regulation of stem cell proliferation(GO:0072091) |
| 0.2 | 3.7 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 2.2 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.2 | 2.9 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.2 | 2.4 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.2 | 5.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.2 | 0.7 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.2 | 8.7 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.2 | 0.8 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 3.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.7 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.1 | 0.8 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.6 | GO:1901909 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 1.7 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.1 | 0.5 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 3.2 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.1 | 0.8 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.3 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 1.2 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 1.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.6 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 0.3 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 2.1 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.1 | 1.9 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 1.3 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 0.7 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 0.6 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.1 | 1.0 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 1.5 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.6 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 1.4 | GO:0072329 | monocarboxylic acid catabolic process(GO:0072329) |
| 0.0 | 2.4 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 1.1 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 1.3 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 1.2 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 2.0 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 1.0 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.6 | GO:0030217 | T cell differentiation(GO:0030217) |
| 0.0 | 0.5 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.9 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.9 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.6 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.3 | 3.8 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.2 | 2.4 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.2 | 3.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 5.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 2.3 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 2.0 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.1 | 0.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 5.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.9 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.9 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 6.2 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 1.5 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.5 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 1.0 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 6.1 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 3.4 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 8.7 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 1.1 | 4.2 | GO:1903231 | mRNA binding involved in posttranscriptional gene silencing(GO:1903231) |
| 0.8 | 3.8 | GO:0009374 | biotin binding(GO:0009374) |
| 0.6 | 3.8 | GO:0090624 | endoribonuclease activity, cleaving miRNA-paired mRNA(GO:0090624) |
| 0.5 | 1.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.5 | 1.4 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.3 | 1.3 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.3 | 2.6 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.2 | 1.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 1.6 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.2 | 1.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 6.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.2 | 2.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 0.7 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.2 | 0.7 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 0.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 4.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 5.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 1.0 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 2.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 2.2 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 1.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 2.2 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.1 | 0.6 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.3 | GO:0035620 | ceramide transporter activity(GO:0035620) sphingolipid transporter activity(GO:0046624) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.1 | 2.3 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 0.7 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.6 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 5.3 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.1 | 1.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.8 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 0.2 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.6 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.6 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 1.9 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.9 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 4.7 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 3.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.4 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 2.4 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.3 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 2.2 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.3 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 1.2 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.0 | 0.8 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 2.1 | GO:0042393 | histone binding(GO:0042393) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.0 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 1.6 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 8.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.6 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 0.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.0 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 2.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.7 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.2 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |