Project
DANIO-CODE
Navigation
Downloads
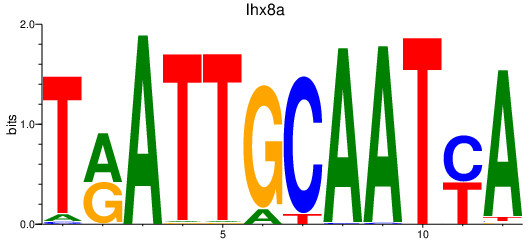
Results for lhx8a
Z-value: 0.64
Transcription factors associated with lhx8a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lhx8a
|
ENSDARG00000002330 | LIM homeobox 8a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lhx8a | dr10_dc_chr2_+_11422195_11422248 | -0.55 | 2.9e-02 | Click! |
Activity profile of lhx8a motif
Sorted Z-values of lhx8a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of lhx8a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_35913418 | 1.42 |
ENSDART00000013590
|
wnt11
|
wingless-type MMTV integration site family, member 11 |
| chr9_-_30560440 | 1.20 |
ENSDART00000079068
|
asb11
|
ankyrin repeat and SOCS box containing 11 |
| chr15_-_14102102 | 0.84 |
ENSDART00000139068
|
zgc:114130
|
zgc:114130 |
| chr6_-_32106882 | 0.74 |
ENSDART00000144772
|
BX664614.1
|
ENSDARG00000095311 |
| chr15_-_643531 | 0.73 |
ENSDART00000102257
|
alox5b.3
|
arachidonate 5-lipoxygenase b, tandem duplicate 3 |
| chr7_-_52850565 | 0.54 |
|
|
|
| chr17_-_22532811 | 0.53 |
ENSDART00000079401
|
si:ch211-125o16.4
|
si:ch211-125o16.4 |
| chr7_+_48597712 | 0.52 |
ENSDART00000009642
|
igf2a
|
insulin-like growth factor 2a |
| chr13_+_29794944 | 0.49 |
ENSDART00000123482
|
cuedc2
|
CUE domain containing 2 |
| KN150334v1_-_9585 | 0.48 |
ENSDART00000175935
|
CABZ01113810.1
|
ENSDARG00000107898 |
| chr21_-_13565401 | 0.43 |
ENSDART00000151547
|
pnpla7a
|
patatin-like phospholipase domain containing 7a |
| chr13_+_29161591 | 0.41 |
ENSDART00000115023
|
PARG
|
poly(ADP-ribose) glycohydrolase |
| chr3_-_21006698 | 0.41 |
ENSDART00000104051
|
cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr5_+_26925238 | 0.39 |
ENSDART00000051491
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr16_-_12582192 | 0.38 |
|
|
|
| chr21_-_34891991 | 0.38 |
ENSDART00000065337
|
kif20a
|
kinesin family member 20A |
| chr19_+_365719 | 0.37 |
ENSDART00000093383
|
vps72
|
vacuolar protein sorting 72 homolog (S. cerevisiae) |
| chr19_+_365553 | 0.37 |
ENSDART00000093383
|
vps72
|
vacuolar protein sorting 72 homolog (S. cerevisiae) |
| chr4_-_12767472 | 0.37 |
ENSDART00000024312
|
dera
|
deoxyribose-phosphate aldolase (putative) |
| chr16_-_25620142 | 0.37 |
ENSDART00000077420
|
dtnbp1a
|
dystrobrevin binding protein 1a |
| chr3_-_15529158 | 0.35 |
ENSDART00000080441
|
zgc:66443
|
zgc:66443 |
| chr3_+_28834898 | 0.34 |
|
|
|
| chr21_-_11877525 | 0.33 |
ENSDART00000145194
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr24_-_23155932 | 0.32 |
|
|
|
| chr19_+_365865 | 0.32 |
ENSDART00000093383
|
vps72
|
vacuolar protein sorting 72 homolog (S. cerevisiae) |
| chr17_-_11312603 | 0.32 |
ENSDART00000091159
|
adpgk2
|
ADP-dependent glucokinase 2 |
| chr5_-_66571831 | 0.31 |
ENSDART00000114783
|
clip1a
|
CAP-GLY domain containing linker protein 1a |
| chr24_+_36451108 | 0.31 |
ENSDART00000142264
|
grnb
|
granulin b |
| chr17_-_29254409 | 0.31 |
ENSDART00000104219
|
rcor1
|
REST corepressor 1 |
| chr8_-_23591293 | 0.31 |
ENSDART00000025024
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr23_+_32174669 | 0.30 |
ENSDART00000000992
|
ENSDARG00000000887
|
ENSDARG00000000887 |
| chr1_-_30299017 | 0.30 |
|
|
|
| chr3_+_28834958 | 0.30 |
|
|
|
| chr19_-_43434166 | 0.30 |
|
|
|
| chr24_-_32110851 | 0.29 |
ENSDART00000159034
|
rsu1
|
Ras suppressor protein 1 |
| chr5_+_26925392 | 0.29 |
ENSDART00000051491
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr8_-_23591082 | 0.28 |
ENSDART00000025024
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr11_+_36093297 | 0.27 |
ENSDART00000077649
|
sypl2a
|
synaptophysin-like 2a |
| chr8_+_23704663 | 0.27 |
ENSDART00000104346
|
mkrn4
|
makorin, ring finger protein, 4 |
| chr15_-_20903489 | 0.27 |
ENSDART00000139551
|
aldh3a2a
|
aldehyde dehydrogenase 3 family, member A2a |
| chr17_-_29253918 | 0.26 |
ENSDART00000104219
|
rcor1
|
REST corepressor 1 |
| chr16_+_28829383 | 0.26 |
ENSDART00000122433
|
trim33l
|
tripartite motif containing 33, like |
| chr24_-_9549374 | 0.26 |
ENSDART00000093046
|
uba5
|
ubiquitin-like modifier activating enzyme 5 |
| chr11_+_25039127 | 0.26 |
ENSDART00000126211
|
cyldb
|
cylindromatosis (turban tumor syndrome), b |
| chr25_-_325847 | 0.26 |
ENSDART00000059514
|
prickle1a
|
prickle homolog 1a |
| chr15_-_20903444 | 0.26 |
ENSDART00000139551
|
aldh3a2a
|
aldehyde dehydrogenase 3 family, member A2a |
| chr25_-_3682900 | 0.25 |
ENSDART00000154691
|
gatd1
|
glutamine amidotransferase like class 1 domain containing 1 |
| chr18_+_20058174 | 0.25 |
ENSDART00000146957
|
uacaa
|
uveal autoantigen with coiled-coil domains and ankyrin repeats a |
| chr20_+_50212438 | 0.25 |
ENSDART00000097729
|
riox1
|
ribosomal oxygenase 1 |
| chr20_+_38955797 | 0.24 |
|
|
|
| chr15_+_22458649 | 0.23 |
ENSDART00000109931
|
oafa
|
OAF homolog a (Drosophila) |
| chr19_+_816354 | 0.23 |
ENSDART00000093304
|
nrm
|
nurim (nuclear envelope membrane protein |
| chr6_+_32107019 | 0.23 |
ENSDART00000155710
|
BX664614.2
|
ENSDARG00000097944 |
| chr6_+_23476669 | 0.23 |
|
|
|
| chr5_+_35913494 | 0.22 |
ENSDART00000013590
|
wnt11
|
wingless-type MMTV integration site family, member 11 |
| chr1_-_6989681 | 0.22 |
ENSDART00000013264
|
arglu1b
|
arginine and glutamate rich 1b |
| chr9_+_46748457 | 0.22 |
|
|
|
| chr18_+_6034458 | 0.22 |
|
|
|
| chr13_+_29794969 | 0.22 |
ENSDART00000123482
|
cuedc2
|
CUE domain containing 2 |
| chr1_-_30271088 | 0.21 |
|
|
|
| chr3_+_10182687 | 0.21 |
ENSDART00000156700
|
cbx2
|
chromobox homolog 2 (Drosophila Pc class) |
| chr20_-_28531019 | 0.21 |
ENSDART00000172133
|
CABZ01057122.1
|
ENSDARG00000105192 |
| chr13_+_8560739 | 0.21 |
ENSDART00000075054
|
thada
|
thyroid adenoma associated |
| chr12_+_19226616 | 0.21 |
ENSDART00000066388
|
tomm22
|
translocase of outer mitochondrial membrane 22 homolog (yeast) |
| chr21_-_39900518 | 0.20 |
ENSDART00000175055
|
CABZ01065291.1
|
ENSDARG00000107374 |
| chr17_-_29254052 | 0.20 |
ENSDART00000104219
|
rcor1
|
REST corepressor 1 |
| chr6_-_31380261 | 0.20 |
|
|
|
| chr4_-_998370 | 0.19 |
|
|
|
| chr15_-_36490729 | 0.19 |
|
|
|
| chr7_+_29969850 | 0.19 |
ENSDART00000109243
|
sema4bb
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Bb |
| chr15_-_35008125 | 0.19 |
ENSDART00000099723
|
sh3bp5la
|
SH3-binding domain protein 5-like, a |
| chr18_+_45199548 | 0.18 |
|
|
|
| chr8_-_5834829 | 0.18 |
ENSDART00000179217
|
ENSDARG00000106522
|
ENSDARG00000106522 |
| chr24_-_9549419 | 0.18 |
ENSDART00000093046
|
uba5
|
ubiquitin-like modifier activating enzyme 5 |
| chr23_-_19125463 | 0.18 |
ENSDART00000111852
|
arfgef2
|
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) |
| chr19_+_44310050 | 0.18 |
ENSDART00000168725
ENSDART00000133628 |
ankib1a
|
ankyrin repeat and IBR domain containing 1a |
| chr1_+_45147775 | 0.18 |
ENSDART00000179047
|
map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr4_-_5766814 | 0.18 |
ENSDART00000021753
|
ccnc
|
cyclin C |
| chr5_+_45823481 | 0.17 |
|
|
|
| chr17_-_29253770 | 0.17 |
ENSDART00000104219
|
rcor1
|
REST corepressor 1 |
| chr16_-_35474091 | 0.17 |
ENSDART00000172294
ENSDART00000162733 |
ctps1b
|
CTP synthase 1b |
| chr20_-_28531087 | 0.16 |
ENSDART00000172133
|
CABZ01057122.1
|
ENSDARG00000105192 |
| chr3_+_33990660 | 0.16 |
ENSDART00000174929
|
aldh3b1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr20_-_33802212 | 0.16 |
ENSDART00000166573
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr6_-_16328987 | 0.16 |
ENSDART00000083305
|
slc19a2
|
solute carrier family 19 (thiamine transporter), member 2 |
| chr16_-_15496868 | 0.16 |
ENSDART00000053754
|
has2
|
hyaluronan synthase 2 |
| chr5_+_36054658 | 0.16 |
ENSDART00000131339
|
capns1a
|
calpain, small subunit 1 a |
| chr17_-_29254289 | 0.16 |
ENSDART00000104219
|
rcor1
|
REST corepressor 1 |
| chr6_+_31381090 | 0.15 |
ENSDART00000038990
|
jak1
|
Janus kinase 1 |
| chr2_+_42155813 | 0.15 |
ENSDART00000143562
|
gbp2
|
guanylate binding protein 2 |
| chr2_+_1260407 | 0.15 |
|
|
|
| chr23_-_18671336 | 0.15 |
ENSDART00000008847
|
ikbkg
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr23_+_29431734 | 0.15 |
ENSDART00000027255
|
tardbpl
|
TAR DNA binding protein, like |
| chr4_+_11480865 | 0.15 |
ENSDART00000019458
|
asb13a.1
|
ankyrin repeat and SOCS box containing 13a, tandem duplicate 1 |
| chr20_-_26517630 | 0.15 |
|
|
|
| chr12_+_47729272 | 0.15 |
ENSDART00000159120
|
polr3a
|
polymerase (RNA) III (DNA directed) polypeptide A |
| chr15_+_22458820 | 0.15 |
ENSDART00000109931
|
oafa
|
OAF homolog a (Drosophila) |
| chr3_-_9755642 | 0.14 |
ENSDART00000168366
|
crebbpb
|
CREB binding protein b |
| chr8_-_16689736 | 0.14 |
ENSDART00000049676
|
depdc1a
|
DEP domain containing 1a |
| chr17_-_50358813 | 0.14 |
|
|
|
| chr23_-_18671263 | 0.14 |
ENSDART00000008847
|
ikbkg
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr3_-_15529108 | 0.14 |
ENSDART00000080441
|
zgc:66443
|
zgc:66443 |
| chr6_+_31381039 | 0.14 |
ENSDART00000038990
|
jak1
|
Janus kinase 1 |
| chr18_-_1631813 | 0.14 |
|
|
|
| chr1_+_29058085 | 0.14 |
ENSDART00000133905
|
pcca
|
propionyl CoA carboxylase, alpha polypeptide |
| chr7_-_39107780 | 0.14 |
ENSDART00000173659
|
slc8b1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr21_+_20350218 | 0.14 |
ENSDART00000144366
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr5_+_61278798 | 0.13 |
ENSDART00000168808
|
si:dkeyp-117b8.4
|
si:dkeyp-117b8.4 |
| chr1_-_7882397 | 0.13 |
ENSDART00000114613
|
ptcd1
|
pentatricopeptide repeat domain 1 |
| chr4_+_26064174 | 0.13 |
ENSDART00000171204
|
SCYL2
|
SCY1 like pseudokinase 2 |
| chr7_+_34923319 | 0.13 |
|
|
|
| chr24_-_8585312 | 0.13 |
|
|
|
| chr18_+_26916897 | 0.13 |
ENSDART00000050230
|
tspan3a
|
tetraspanin 3a |
| KN150702v1_-_111618 | 0.13 |
ENSDART00000165431
|
PPP1CC
|
protein phosphatase 1 catalytic subunit gamma |
| chr9_-_30560596 | 0.13 |
ENSDART00000079068
|
asb11
|
ankyrin repeat and SOCS box containing 11 |
| chr18_+_38918883 | 0.13 |
ENSDART00000159202
|
myo5aa
|
myosin VAa |
| chr3_+_19536018 | 0.12 |
ENSDART00000006490
|
tlk2
|
tousled-like kinase 2 |
| chr19_-_12032026 | 0.12 |
ENSDART00000024861
ENSDART00000110513 ENSDART00000163478 |
cpsf1
|
cleavage and polyadenylation specific factor 1 |
| chr16_+_28829592 | 0.12 |
ENSDART00000122433
|
trim33l
|
tripartite motif containing 33, like |
| chr21_+_9904675 | 0.12 |
ENSDART00000171579
|
herc7
|
hect domain and RLD 7 |
| chr12_-_3672263 | 0.12 |
|
|
|
| chr17_-_51032797 | 0.11 |
ENSDART00000130412
ENSDART00000123746 ENSDART00000162717 |
aqr
|
aquarius intron-binding spliceosomal factor |
| chr13_-_30031075 | 0.11 |
ENSDART00000040409
|
ppa1b
|
pyrophosphatase (inorganic) 1b |
| chr16_-_37025649 | 0.11 |
ENSDART00000113834
|
snrnp48
|
small nuclear ribonucleoprotein 48 (U11/U12) |
| chr17_+_24571678 | 0.11 |
ENSDART00000092941
|
rlf
|
rearranged L-myc fusion |
| chr1_-_6989807 | 0.11 |
ENSDART00000013264
|
arglu1b
|
arginine and glutamate rich 1b |
| chr7_-_39107722 | 0.11 |
ENSDART00000173659
|
slc8b1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr7_-_59851104 | 0.11 |
ENSDART00000128363
|
cct7
|
chaperonin containing TCP1, subunit 7 (eta) |
| chr12_+_19226572 | 0.11 |
ENSDART00000066388
|
tomm22
|
translocase of outer mitochondrial membrane 22 homolog (yeast) |
| chr21_+_9904707 | 0.11 |
ENSDART00000171579
|
herc7
|
hect domain and RLD 7 |
| chr21_-_3299950 | 0.10 |
|
|
|
| chr1_+_43471683 | 0.10 |
ENSDART00000166324
|
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr5_-_40428786 | 0.10 |
|
|
|
| chr13_-_8560620 | 0.10 |
ENSDART00000144553
|
plekhh2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2 |
| chr15_+_25747735 | 0.10 |
ENSDART00000148190
|
hic1
|
hypermethylated in cancer 1 |
| chr25_+_19140255 | 0.10 |
ENSDART00000110730
|
ppip5k1b
|
diphosphoinositol pentakisphosphate kinase 1b |
| chr8_+_26336302 | 0.10 |
|
|
|
| chr10_+_17480699 | 0.10 |
|
|
|
| chr21_-_16980069 | 0.09 |
|
|
|
| chr9_-_30560373 | 0.09 |
ENSDART00000079068
|
asb11
|
ankyrin repeat and SOCS box containing 11 |
| chr7_+_22552724 | 0.09 |
ENSDART00000101459
ENSDART00000159743 |
pygmb
|
phosphorylase, glycogen, muscle b |
| chr16_+_28829508 | 0.09 |
ENSDART00000122433
|
trim33l
|
tripartite motif containing 33, like |
| chr23_+_43916552 | 0.09 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr9_-_4640088 | 0.09 |
ENSDART00000179110
|
galnt13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 |
| chr15_-_47351885 | 0.09 |
|
|
|
| chr12_-_17741310 | 0.09 |
ENSDART00000166604
|
baiap2l1a
|
BAI1-associated protein 2-like 1a |
| chr20_+_6545449 | 0.09 |
ENSDART00000145763
|
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr21_+_15505169 | 0.09 |
ENSDART00000011318
|
smarcb1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1b |
| chr24_-_7603837 | 0.08 |
|
|
|
| chr1_-_57300743 | 0.08 |
ENSDART00000081122
|
COLGALT1 (1 of many)
|
collagen beta(1-O)galactosyltransferase 1 |
| chr23_+_43916520 | 0.08 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr17_-_29254184 | 0.08 |
ENSDART00000104219
|
rcor1
|
REST corepressor 1 |
| chr1_+_54417596 | 0.08 |
ENSDART00000132727
|
aftpha
|
aftiphilin a |
| chr21_+_27266000 | 0.08 |
ENSDART00000012855
|
sart1
|
squamous cell carcinoma antigen recognised by T cells |
| chr22_-_7887401 | 0.08 |
ENSDART00000097198
|
sc:d217
|
sc:d217 |
| chr5_-_14993912 | 0.08 |
ENSDART00000085943
|
taok3a
|
TAO kinase 3a |
| chr4_-_5083532 | 0.07 |
ENSDART00000130453
|
ahcyl2
|
adenosylhomocysteinase-like 2 |
| chr18_-_12484760 | 0.07 |
ENSDART00000135574
|
AL929229.1
|
ENSDARG00000091975 |
| chr14_-_6669062 | 0.07 |
|
|
|
| chr18_-_21116872 | 0.07 |
ENSDART00000100791
|
si:dkey-12e7.1
|
si:dkey-12e7.1 |
| chr7_+_20651600 | 0.07 |
ENSDART00000129161
|
wrap53
|
WD repeat containing, antisense to TP53 |
| chr14_-_33605295 | 0.07 |
ENSDART00000168546
|
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr19_+_27055869 | 0.07 |
ENSDART00000067793
ENSDART00000124755 |
ints3
|
integrator complex subunit 3 |
| chr20_-_873476 | 0.07 |
ENSDART00000142361
ENSDART00000104725 |
snx14
|
sorting nexin 14 |
| chr6_-_53333532 | 0.07 |
ENSDART00000172465
|
gnb1b
|
guanine nucleotide binding protein (G protein), beta polypeptide 1b |
| chr10_-_7927246 | 0.07 |
|
|
|
| chr23_-_27768575 | 0.06 |
ENSDART00000146703
|
IKZF4
|
IKAROS family zinc finger 4 |
| chr9_+_28301359 | 0.06 |
|
|
|
| chr9_+_28301214 | 0.06 |
|
|
|
| chr23_-_27647510 | 0.06 |
|
|
|
| chr16_-_23975775 | 0.06 |
ENSDART00000077807
|
BX004983.1
|
ENSDARG00000055446 |
| chr18_-_26830520 | 0.06 |
ENSDART00000161281
|
znf592
|
zinc finger protein 592 |
| chr15_+_29460803 | 0.06 |
ENSDART00000155198
|
gdpd5b
|
glycerophosphodiester phosphodiesterase domain containing 5b |
| chr23_+_21352308 | 0.06 |
ENSDART00000033970
|
ubr4
|
ubiquitin protein ligase E3 component n-recognin 4 |
| chr18_-_7580225 | 0.06 |
ENSDART00000133541
|
si:dkey-30c15.2
|
si:dkey-30c15.2 |
| chr7_+_16257193 | 0.06 |
ENSDART00000160263
|
zdhhc13
|
zinc finger, DHHC-type containing 13 |
| chr12_-_34614903 | 0.06 |
ENSDART00000152876
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr1_+_29058115 | 0.05 |
ENSDART00000135252
|
pcca
|
propionyl CoA carboxylase, alpha polypeptide |
| chr23_-_1631192 | 0.05 |
|
|
|
| chr21_-_2292663 | 0.05 |
ENSDART00000164015
|
zgc:66483
|
zgc:66483 |
| chr16_+_28830019 | 0.05 |
|
|
|
| chr6_+_11014565 | 0.05 |
ENSDART00000132677
|
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr25_+_21001016 | 0.05 |
ENSDART00000079012
|
rad52
|
RAD52 homolog, DNA repair protein |
| chr18_-_13088412 | 0.05 |
|
|
|
| chr12_+_47729417 | 0.05 |
ENSDART00000159120
|
polr3a
|
polymerase (RNA) III (DNA directed) polypeptide A |
| chr21_-_16979722 | 0.05 |
|
|
|
| chr1_-_2309740 | 0.05 |
ENSDART00000103795
|
ggact.1
|
gamma-glutamylamine cyclotransferase, tandem duplicate 1 |
| chr9_-_8035360 | 0.05 |
|
|
|
| chr15_-_3921649 | 0.05 |
ENSDART00000157583
|
rnf168
|
ring finger protein 168 |
| chr18_-_31126989 | 0.05 |
ENSDART00000039495
|
pdcd5
|
programmed cell death 5 |
| chr4_-_28190966 | 0.04 |
|
|
|
| chr3_+_15244806 | 0.04 |
ENSDART00000133168
|
atxn2l
|
ataxin 2-like |
| chr20_-_29630048 | 0.04 |
ENSDART00000049224
|
taf1b
|
TATA box binding protein (Tbp)-associated factor, RNA polymerase I, B |
| chr22_-_15576405 | 0.04 |
ENSDART00000009054
|
tpm4a
|
tropomyosin 4a |
| chr3_+_25036265 | 0.04 |
ENSDART00000077493
|
TST
|
thiosulfate sulfurtransferase |
| chr1_-_20845715 | 0.04 |
ENSDART00000131257
|
wdr19
|
WD repeat domain 19 |
| chr13_-_24130160 | 0.04 |
ENSDART00000138747
ENSDART00000101168 |
urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr5_-_8260713 | 0.04 |
ENSDART00000167793
|
mybbp1a
|
MYB binding protein (P160) 1a |
| chr20_-_51915872 | 0.03 |
ENSDART00000004092
|
brox
|
BRO1 domain and CAAX motif containing |
| chr22_-_11565090 | 0.03 |
ENSDART00000137537
|
gnav1
|
guanine nucleotide binding protein (G protein) alpha v1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.2 | 0.6 | GO:0032329 | serine transport(GO:0032329) L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.2 | 0.7 | GO:0051121 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.2 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 1.2 | GO:0030656 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.1 | 0.4 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.1 | 0.3 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 0.2 | GO:0003091 | renal water homeostasis(GO:0003091) renal water transport(GO:0003097) extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.3 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 | 0.4 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.5 | GO:0070665 | positive regulation of mononuclear cell proliferation(GO:0032946) positive regulation of T cell proliferation(GO:0042102) positive regulation of lymphocyte proliferation(GO:0050671) positive regulation of leukocyte proliferation(GO:0070665) |
| 0.0 | 0.4 | GO:0009264 | deoxyribonucleotide catabolic process(GO:0009264) deoxyribose phosphate catabolic process(GO:0046386) |
| 0.0 | 0.4 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.2 | GO:0045117 | azole transport(GO:0045117) |
| 0.0 | 0.3 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.3 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 1.6 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.1 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.7 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.3 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.2 | GO:0019856 | pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.2 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.1 | GO:0010668 | ectodermal cell fate commitment(GO:0001712) ectodermal cell fate specification(GO:0001715) ectodermal cell differentiation(GO:0010668) regulation of ectodermal cell fate specification(GO:0042665) regulation of ectoderm development(GO:2000383) |
| 0.0 | 0.2 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.6 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 0.1 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.0 | 0.0 | GO:0002312 | B cell activation involved in immune response(GO:0002312) isotype switching(GO:0045190) |
| 0.0 | 0.2 | GO:0002042 | cell migration involved in sprouting angiogenesis(GO:0002042) |
| 0.0 | 0.1 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.1 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.4 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 1.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.0 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.2 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 0.2 | GO:1901474 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.1 | 0.7 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 0.5 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.6 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 1.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.7 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.2 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.3 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.2 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.4 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.7 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.3 | GO:0030515 | snoRNA binding(GO:0030515) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.5 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 1.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.2 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.2 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |