Project
DANIO-CODE
Navigation
Downloads
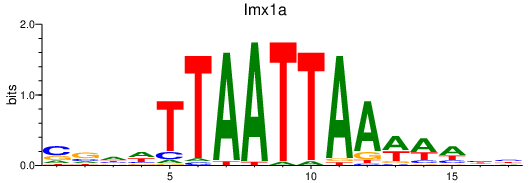
Results for lmx1a
Z-value: 0.54
Transcription factors associated with lmx1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lmx1a
|
ENSDARG00000020354 | LIM homeobox transcription factor 1, alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lmx1a | dr10_dc_chr20_+_34021116_34021138 | -0.77 | 4.3e-04 | Click! |
Activity profile of lmx1a motif
Sorted Z-values of lmx1a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of lmx1a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_782643 | 1.57 |
ENSDART00000161084
|
si:dkey-205h23.1
|
si:dkey-205h23.1 |
| chr12_+_25509394 | 1.44 |
ENSDART00000077157
|
six3b
|
SIX homeobox 3b |
| chr8_-_26623251 | 1.42 |
ENSDART00000159264
|
sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr7_+_6814828 | 1.35 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr20_-_45908435 | 1.32 |
ENSDART00000147637
|
fermt1
|
fermitin family member 1 |
| chr14_-_17257773 | 1.26 |
ENSDART00000082667
|
fgfrl1a
|
fibroblast growth factor receptor-like 1a |
| chr20_-_9107294 | 1.09 |
ENSDART00000140792
|
OMA1
|
OMA1 zinc metallopeptidase |
| chr3_+_49277125 | 1.03 |
ENSDART00000161724
|
gas7a
|
growth arrest-specific 7a |
| chr1_+_13779755 | 1.02 |
|
|
|
| chr2_-_49114158 | 0.98 |
|
|
|
| chr21_-_1716495 | 0.89 |
ENSDART00000151049
|
onecut2
|
one cut homeobox 2 |
| chr7_+_25052687 | 0.79 |
ENSDART00000110347
|
cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr6_+_8895437 | 0.79 |
ENSDART00000162588
|
rgn
|
regucalcin |
| chr18_+_46384462 | 0.75 |
ENSDART00000024202
|
daw1
|
dynein assembly factor with WDR repeat domains 1 |
| chr17_+_30352361 | 0.75 |
ENSDART00000076611
|
greb1
|
growth regulation by estrogen in breast cancer 1 |
| chr6_-_43679553 | 0.71 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr16_-_16244210 | 0.64 |
ENSDART00000012718
|
fabp11b
|
fatty acid binding protein 11b |
| chr1_-_674449 | 0.62 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr19_+_5562107 | 0.62 |
ENSDART00000082080
|
jupb
|
junction plakoglobin b |
| chr5_-_31301280 | 0.61 |
ENSDART00000141446
|
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr13_-_29290894 | 0.58 |
ENSDART00000150228
|
chata
|
choline O-acetyltransferase a |
| chr16_-_28921444 | 0.52 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr22_-_12130441 | 0.50 |
ENSDART00000146785
|
tmem163b
|
transmembrane protein 163b |
| chr9_+_34832049 | 0.50 |
ENSDART00000100735
ENSDART00000133996 |
shox
|
short stature homeobox |
| chr18_-_605558 | 0.49 |
ENSDART00000157564
|
wtip
|
WT1 interacting protein |
| chr2_-_30005139 | 0.49 |
ENSDART00000146968
|
cnpy1
|
canopy1 |
| chr9_-_43736549 | 0.49 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr8_+_43334181 | 0.48 |
ENSDART00000038566
|
fam101a
|
family with sequence similarity 101, member A |
| chr4_-_20091845 | 0.47 |
|
|
|
| chr10_+_11303321 | 0.45 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr12_+_18559530 | 0.44 |
ENSDART00000152948
|
rgs9b
|
regulator of G protein signaling 9b |
| chr24_+_24316486 | 0.44 |
ENSDART00000147658
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr7_+_21006803 | 0.43 |
ENSDART00000052942
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr22_-_12905648 | 0.43 |
ENSDART00000157820
|
mfsd6a
|
major facilitator superfamily domain containing 6a |
| chr18_-_43890514 | 0.42 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr14_-_14302241 | 0.41 |
ENSDART00000162322
|
rab9b
|
RAB9B, member RAS oncogene family |
| chr21_-_5692160 | 0.40 |
ENSDART00000075137
ENSDART00000151202 |
ccni
|
cyclin I |
| chr17_+_25425967 | 0.40 |
ENSDART00000030691
|
clic4
|
chloride intracellular channel 4 |
| chr5_+_45823481 | 0.38 |
|
|
|
| chr1_-_9256864 | 0.37 |
ENSDART00000041849
|
tmem8a
|
transmembrane protein 8A |
| chr8_-_12253026 | 0.35 |
ENSDART00000091612
|
dab2ipa
|
DAB2 interacting protein a |
| chr8_-_14446883 | 0.32 |
ENSDART00000057644
|
lhx4
|
LIM homeobox 4 |
| chr13_+_32609784 | 0.30 |
ENSDART00000160138
|
sobpa
|
sine oculis binding protein homolog (Drosophila) a |
| chr3_+_27667194 | 0.29 |
ENSDART00000075100
|
carhsp1
|
calcium regulated heat stable protein 1 |
| chr15_+_32940372 | 0.24 |
ENSDART00000170098
|
spg20b
|
spastic paraplegia 20b (Troyer syndrome) |
| chr24_+_24316408 | 0.22 |
ENSDART00000147658
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr19_-_46058123 | 0.22 |
|
|
|
| chr21_+_42893361 | 0.20 |
|
|
|
| chr13_+_32609849 | 0.20 |
ENSDART00000160138
|
sobpa
|
sine oculis binding protein homolog (Drosophila) a |
| chr2_-_16548451 | 0.17 |
ENSDART00000152031
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr7_+_7463292 | 0.16 |
ENSDART00000091099
|
ino80b
|
INO80 complex subunit B |
| chr24_+_24316346 | 0.13 |
ENSDART00000147658
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr1_+_9634016 | 0.10 |
ENSDART00000029774
|
tmem55bb
|
transmembrane protein 55Bb |
| chr2_+_1881022 | 0.10 |
ENSDART00000101038
|
tmie
|
transmembrane inner ear |
| chr23_+_17952613 | 0.09 |
ENSDART00000156331
|
si:ch73-390p7.2
|
si:ch73-390p7.2 |
| chr11_-_23333850 | 0.08 |
|
|
|
| chr16_+_2582656 | 0.06 |
ENSDART00000092299
|
ENSDARG00000073784
|
ENSDARG00000073784 |
| chr18_+_1353857 | 0.06 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr7_+_58427742 | 0.06 |
ENSDART00000073640
|
plag1
|
pleiomorphic adenoma gene 1 |
| chr18_+_8362131 | 0.04 |
ENSDART00000092053
|
chkb
|
choline kinase beta |
| chr6_+_40924749 | 0.03 |
ENSDART00000133599
|
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr1_+_9633924 | 0.02 |
ENSDART00000029774
|
tmem55bb
|
transmembrane protein 55Bb |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.2 | 0.8 | GO:0042364 | L-ascorbic acid metabolic process(GO:0019852) water-soluble vitamin biosynthetic process(GO:0042364) regulation of calcium-mediated signaling(GO:0050848) |
| 0.1 | 0.4 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 1.3 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.1 | 0.6 | GO:0008291 | neuromuscular synaptic transmission(GO:0007274) acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.1 | 1.4 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.1 | 0.2 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.1 | 0.3 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 0.7 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.1 | 0.3 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.7 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 0.5 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 1.4 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.4 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 1.3 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.5 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.5 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.5 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.1 | GO:0060088 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 0.5 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.3 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.0 | GO:0005884 | actin filament(GO:0005884) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 1.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.6 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 1.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |