Project
DANIO-CODE
Navigation
Downloads
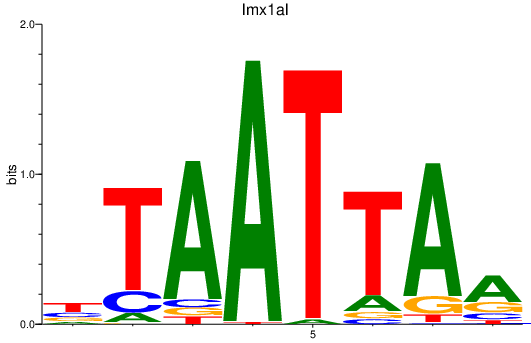
Results for lmx1al
Z-value: 0.78
Transcription factors associated with lmx1al
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lmx1al
|
ENSDARG00000077915 | LIM homeobox transcription factor 1, alpha-like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lmx1al | dr10_dc_chr3_+_52551672_52551677 | 0.28 | 3.0e-01 | Click! |
Activity profile of lmx1al motif
Sorted Z-values of lmx1al motif
Network of associatons between targets according to the STRING database.
First level regulatory network of lmx1al
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_23516127 | 2.47 |
ENSDART00000004679
|
icn
|
ictacalcin |
| chr20_-_22576513 | 1.96 |
ENSDART00000103510
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr9_-_30437160 | 1.87 |
ENSDART00000147241
|
BX936337.1
|
ENSDARG00000092870 |
| chr24_-_21778717 | 1.82 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr1_+_13779755 | 1.75 |
|
|
|
| chr18_-_3056732 | 1.74 |
ENSDART00000162657
|
rps3
|
ribosomal protein S3 |
| chr9_+_25966093 | 1.67 |
ENSDART00000146011
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr6_+_56157608 | 1.55 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr9_-_41608298 | 1.54 |
|
|
|
| chr2_+_33343287 | 1.42 |
ENSDART00000056655
|
klf17
|
Kruppel-like factor 17 |
| chr1_-_4757890 | 1.40 |
ENSDART00000114035
|
mnx2b
|
motor neuron and pancreas homeobox 2b |
| chr7_+_19300351 | 1.39 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr8_-_22945616 | 1.38 |
|
|
|
| chr25_-_14472107 | 1.34 |
|
|
|
| chr13_+_27102308 | 1.34 |
ENSDART00000145901
|
rin2
|
Ras and Rab interactor 2 |
| chr14_+_46419051 | 1.31 |
ENSDART00000112377
|
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr24_-_14447773 | 1.29 |
|
|
|
| chr3_-_35928594 | 1.29 |
|
|
|
| chr11_+_34788205 | 1.26 |
ENSDART00000124800
|
fam212aa
|
family with sequence similarity 212, member Aa |
| chr19_+_7234029 | 1.22 |
ENSDART00000080348
|
brd2a
|
bromodomain containing 2a |
| chr8_-_26623251 | 1.22 |
ENSDART00000159264
|
sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr21_+_26684617 | 1.19 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr19_+_14197118 | 1.16 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr13_+_27102377 | 1.15 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr16_-_42990753 | 1.11 |
ENSDART00000149317
|
hfe2
|
hemochromatosis type 2 |
| chr9_-_12453581 | 1.11 |
ENSDART00000088199
|
zgc:162707
|
zgc:162707 |
| chr5_-_40707316 | 1.10 |
ENSDART00000161932
|
npr3
|
natriuretic peptide receptor 3 |
| chr1_+_32935645 | 1.10 |
ENSDART00000170832
|
arl13b
|
ADP-ribosylation factor-like 13b |
| chr6_-_18891908 | 1.09 |
ENSDART00000074327
|
igfbp2a
|
insulin-like growth factor binding protein 2a |
| chr9_-_20562293 | 1.09 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr15_-_40391882 | 1.09 |
|
|
|
| chr13_+_35213326 | 1.08 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr20_-_475417 | 1.08 |
ENSDART00000032212
|
fynrk
|
fyn-related Src family tyrosine kinase |
| chr9_-_48098629 | 1.08 |
|
|
|
| chr24_-_6048914 | 1.06 |
ENSDART00000146830
ENSDART00000021981 |
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr3_-_6078015 | 1.03 |
ENSDART00000165715
|
BX284638.1
|
ENSDARG00000098850 |
| chr23_+_11350727 | 1.02 |
|
|
|
| chr4_+_16896821 | 1.00 |
ENSDART00000017726
|
etnk1
|
ethanolamine kinase 1 |
| chr11_-_41241693 | 0.99 |
ENSDART00000002556
|
mrto4
|
MRT4 homolog, ribosome maturation factor |
| chr17_+_23278879 | 0.99 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr8_+_21321765 | 0.97 |
ENSDART00000131691
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr7_+_6814828 | 0.97 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr14_-_17257773 | 0.96 |
ENSDART00000082667
|
fgfrl1a
|
fibroblast growth factor receptor-like 1a |
| chr16_-_29593569 | 0.96 |
ENSDART00000150028
|
onecutl
|
one cut domain, family member, like |
| chr15_-_14616083 | 0.95 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr6_-_43094573 | 0.94 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr4_-_15442828 | 0.94 |
ENSDART00000157414
|
plxna4
|
plexin A4 |
| chr16_-_17439735 | 0.93 |
ENSDART00000144392
|
zyx
|
zyxin |
| chr6_+_13833991 | 0.91 |
|
|
|
| chr3_+_25992836 | 0.90 |
ENSDART00000010477
|
hsp70.3
|
heat shock cognate 70-kd protein, tandem duplicate 3 |
| chr17_-_2513630 | 0.90 |
ENSDART00000135374
|
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr9_+_25966225 | 0.89 |
ENSDART00000146011
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr16_+_3090170 | 0.89 |
ENSDART00000110395
|
limd1a
|
LIM domains containing 1a |
| chr10_-_11303185 | 0.89 |
ENSDART00000146727
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr2_+_49358871 | 0.89 |
ENSDART00000179089
|
sema6ba
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Ba |
| chr18_-_43890514 | 0.88 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr20_-_9107294 | 0.87 |
ENSDART00000140792
|
OMA1
|
OMA1 zinc metallopeptidase |
| chr19_+_31046291 | 0.87 |
ENSDART00000052124
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr1_-_40536800 | 0.86 |
ENSDART00000134739
|
rgs12b
|
regulator of G protein signaling 12b |
| chr6_+_23787804 | 0.85 |
ENSDART00000163188
|
znf648
|
zinc finger protein 648 |
| chr16_+_24046472 | 0.84 |
|
|
|
| chr2_-_7375138 | 0.82 |
ENSDART00000146434
|
zgc:153115
|
zgc:153115 |
| chr7_+_20251345 | 0.80 |
ENSDART00000157699
|
si:dkey-19b23.12
|
si:dkey-19b23.12 |
| chr20_+_51385187 | 0.79 |
|
|
|
| chr4_+_3970874 | 0.79 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr5_-_40894631 | 0.78 |
ENSDART00000121840
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr15_-_4537178 | 0.78 |
ENSDART00000155619
ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr3_-_60929921 | 0.77 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr23_-_45927723 | 0.76 |
|
|
|
| chr17_+_30352361 | 0.75 |
ENSDART00000076611
|
greb1
|
growth regulation by estrogen in breast cancer 1 |
| chr6_-_43273456 | 0.75 |
|
|
|
| chr17_-_29102320 | 0.74 |
ENSDART00000104204
|
foxg1a
|
forkhead box G1a |
| chr16_+_13928844 | 0.74 |
ENSDART00000090191
|
flcn
|
folliculin |
| chr12_+_24221087 | 0.72 |
ENSDART00000088178
|
nrxn1a
|
neurexin 1a |
| chr18_-_19467100 | 0.72 |
ENSDART00000060363
|
rpl4
|
ribosomal protein L4 |
| chr24_+_21395671 | 0.72 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr17_-_26849495 | 0.72 |
ENSDART00000153590
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr16_+_23172295 | 0.71 |
ENSDART00000167518
ENSDART00000161087 |
efna3b
|
ephrin-A3b |
| chr6_-_23193752 | 0.71 |
ENSDART00000159749
|
usp43a
|
ubiquitin specific peptidase 43a |
| chr6_-_54816183 | 0.70 |
ENSDART00000148462
|
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr20_-_37587804 | 0.69 |
ENSDART00000166106
|
si:ch211-202p1.5
|
si:ch211-202p1.5 |
| chr22_+_16509286 | 0.69 |
ENSDART00000083063
|
tal1
|
T-cell acute lymphocytic leukemia 1 |
| chr20_-_21031504 | 0.67 |
ENSDART00000152726
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr14_+_34151963 | 0.67 |
ENSDART00000144301
|
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr2_+_21267793 | 0.66 |
ENSDART00000099913
|
pla2g4aa
|
phospholipase A2, group IVAa (cytosolic, calcium-dependent) |
| chr17_+_15425559 | 0.66 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr21_-_20674965 | 0.66 |
ENSDART00000065649
|
ENSDARG00000044676
|
ENSDARG00000044676 |
| chr3_+_17387551 | 0.66 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr2_+_3370130 | 0.64 |
ENSDART00000098391
|
wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr20_+_29307039 | 0.64 |
ENSDART00000152949
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr3_-_56474209 | 0.64 |
ENSDART00000156398
|
si:ch211-189a21.1
|
si:ch211-189a21.1 |
| chr20_+_29307142 | 0.61 |
ENSDART00000153016
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr16_+_46145286 | 0.61 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr5_+_18826505 | 0.61 |
|
|
|
| chr3_-_39554155 | 0.61 |
ENSDART00000015393
|
b9d1
|
B9 protein domain 1 |
| chr2_+_50873893 | 0.60 |
|
|
|
| chr14_-_1342450 | 0.58 |
ENSDART00000060417
|
cetn4
|
centrin 4 |
| chr1_-_25600988 | 0.58 |
ENSDART00000160381
|
cxxc4
|
CXXC finger 4 |
| chr5_+_18827478 | 0.58 |
ENSDART00000089078
|
acacb
|
acetyl-CoA carboxylase beta |
| chr1_-_15972040 | 0.57 |
ENSDART00000040434
|
asah1b
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1b |
| chr22_-_7371498 | 0.57 |
|
|
|
| chr25_+_14411153 | 0.56 |
ENSDART00000015681
|
dbx1b
|
developing brain homeobox 1b |
| chr3_-_60930025 | 0.56 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr24_+_35899507 | 0.56 |
ENSDART00000122408
|
si:dkeyp-7a3.1
|
si:dkeyp-7a3.1 |
| chr23_+_31986806 | 0.56 |
ENSDART00000136910
|
armc1l
|
armadillo repeat containing 1, like |
| chr14_+_21816442 | 0.56 |
ENSDART00000109759
|
tmx2a
|
thioredoxin-related transmembrane protein 2a |
| chr6_+_28215039 | 0.54 |
ENSDART00000104394
|
smx5
|
smx5 |
| chr8_-_19166630 | 0.53 |
|
|
|
| chr2_-_49114158 | 0.53 |
|
|
|
| chr1_-_40208544 | 0.52 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr7_-_27877036 | 0.52 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr25_+_4918339 | 0.52 |
ENSDART00000153980
|
parvb
|
parvin, beta |
| chr13_+_28617462 | 0.52 |
ENSDART00000101633
|
prom2
|
prominin 2 |
| chr25_+_24193604 | 0.52 |
ENSDART00000083407
|
b4galnt4a
|
beta-1,4-N-acetyl-galactosaminyl transferase 4a |
| chr14_-_6901415 | 0.51 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr15_-_23441268 | 0.51 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr10_+_11303321 | 0.50 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr16_+_51319421 | 0.50 |
|
|
|
| chr10_+_11809550 | 0.50 |
ENSDART00000092047
|
ppwd1
|
peptidylprolyl isomerase domain and WD repeat containing 1 |
| chr6_-_43094926 | 0.50 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr21_+_28921734 | 0.50 |
ENSDART00000166575
|
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr20_-_28739099 | 0.49 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr2_-_57244462 | 0.49 |
ENSDART00000149235
|
tcf3a
|
transcription factor 3a |
| chr22_+_1274718 | 0.48 |
ENSDART00000159296
|
si:ch73-138e16.4
|
si:ch73-138e16.4 |
| chr9_-_19154264 | 0.48 |
ENSDART00000081878
|
pou1f1
|
POU class 1 homeobox 1 |
| chr16_+_43236190 | 0.48 |
ENSDART00000065643
|
dbf4
|
DBF4 zinc finger |
| chr20_-_29028920 | 0.48 |
ENSDART00000153082
|
susd6
|
sushi domain containing 6 |
| chr7_+_7463292 | 0.48 |
ENSDART00000091099
|
ino80b
|
INO80 complex subunit B |
| chr7_+_24762755 | 0.47 |
ENSDART00000170873
|
sb:cb1058
|
sb:cb1058 |
| chr1_-_50215233 | 0.47 |
ENSDART00000137648
|
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr24_-_24306469 | 0.46 |
ENSDART00000154149
|
BX323067.1
|
ENSDARG00000097984 |
| chr16_+_13993746 | 0.46 |
ENSDART00000101304
|
atg12
|
ATG12 autophagy related 12 homolog (S. cerevisiae) |
| chr8_+_6533379 | 0.46 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr17_+_10437343 | 0.46 |
ENSDART00000167188
|
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr21_-_44707326 | 0.46 |
ENSDART00000013814
|
ammecr1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 homolog (human) |
| chr22_-_12130441 | 0.45 |
ENSDART00000146785
|
tmem163b
|
transmembrane protein 163b |
| chr7_-_6309265 | 0.45 |
ENSDART00000172825
|
FP325123.5
|
Histone H3.2 |
| chr11_-_38282978 | 0.44 |
|
|
|
| chr22_-_12905648 | 0.43 |
ENSDART00000157820
|
mfsd6a
|
major facilitator superfamily domain containing 6a |
| chr20_-_3970778 | 0.42 |
ENSDART00000178724
ENSDART00000178565 |
TRIM67
|
tripartite motif containing 67 |
| chr2_-_36058327 | 0.42 |
ENSDART00000003550
|
nmnat2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr1_-_40208469 | 0.40 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr5_+_35815450 | 0.40 |
ENSDART00000084464
|
fam155b
|
family with sequence similarity 155, member B |
| chr24_+_9272045 | 0.40 |
ENSDART00000132724
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr10_-_32550379 | 0.39 |
ENSDART00000028161
|
uvrag
|
UV radiation resistance associated gene |
| chr22_+_18228143 | 0.39 |
ENSDART00000141535
|
BX664610.1
|
ENSDARG00000095557 |
| chr7_-_26331734 | 0.38 |
|
|
|
| chr22_-_13018196 | 0.38 |
ENSDART00000028787
|
ahr1b
|
aryl hydrocarbon receptor 1b |
| chr3_-_19913881 | 0.38 |
ENSDART00000126915
|
ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr15_+_32963784 | 0.37 |
ENSDART00000167739
|
dclk1b
|
doublecortin-like kinase 1b |
| chr15_+_32963516 | 0.37 |
ENSDART00000167739
|
dclk1b
|
doublecortin-like kinase 1b |
| chr17_-_49355712 | 0.36 |
ENSDART00000004424
|
znf292a
|
zinc finger protein 292a |
| chr5_-_40894693 | 0.36 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr20_-_48677794 | 0.36 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr19_-_42821964 | 0.36 |
ENSDART00000087002
|
plekho1a
|
pleckstrin homology domain containing, family O member 1a |
| chr8_-_18502159 | 0.35 |
ENSDART00000148802
ENSDART00000149081 ENSDART00000148962 |
nexn
|
nexilin (F actin binding protein) |
| chr9_-_43736549 | 0.35 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr6_+_33896684 | 0.35 |
ENSDART00000165710
|
gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr9_+_4708975 | 0.35 |
ENSDART00000081326
|
prpf40a
|
PRP40 pre-mRNA processing factor 40 homolog A |
| chr11_+_36983626 | 0.35 |
ENSDART00000170209
|
il17rc
|
interleukin 17 receptor C |
| chr22_+_38084309 | 0.34 |
ENSDART00000012212
|
commd2
|
COMM domain containing 2 |
| chr5_+_19153421 | 0.34 |
|
|
|
| chr3_+_27667194 | 0.33 |
ENSDART00000075100
|
carhsp1
|
calcium regulated heat stable protein 1 |
| chr16_+_13928376 | 0.33 |
ENSDART00000163251
|
flcn
|
folliculin |
| chr11_+_18710724 | 0.32 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr8_-_11512545 | 0.32 |
ENSDART00000133932
|
si:ch211-248e11.2
|
si:ch211-248e11.2 |
| chr23_+_36016450 | 0.32 |
ENSDART00000103035
|
hoxc6a
|
homeobox C6a |
| chr19_-_10324632 | 0.31 |
ENSDART00000148073
|
shisa7b
|
shisa family member 7 |
| chr6_-_57542101 | 0.31 |
ENSDART00000156967
|
itcha
|
itchy E3 ubiquitin protein ligase a |
| chr8_+_21974863 | 0.31 |
|
|
|
| chr20_-_475321 | 0.30 |
ENSDART00000032212
|
fynrk
|
fyn-related Src family tyrosine kinase |
| chr9_+_36316158 | 0.29 |
ENSDART00000176763
|
lrp1bb
|
low density lipoprotein receptor-related protein 1Bb |
| chr7_+_9726412 | 0.29 |
ENSDART00000173155
|
adamts17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr24_+_14095643 | 0.29 |
ENSDART00000124740
|
ncoa2
|
nuclear receptor coactivator 2 |
| chr7_+_28896401 | 0.29 |
ENSDART00000076345
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr1_+_53384116 | 0.29 |
ENSDART00000149760
|
triobpa
|
TRIO and F-actin binding protein a |
| chr12_+_18559530 | 0.28 |
ENSDART00000152948
|
rgs9b
|
regulator of G protein signaling 9b |
| chr20_-_23526954 | 0.28 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr5_-_67233396 | 0.28 |
ENSDART00000051833
ENSDART00000124890 |
gsx1
|
GS homeobox 1 |
| chr24_+_16402613 | 0.28 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr20_-_46458839 | 0.28 |
ENSDART00000153087
|
bmf2
|
Bcl2 modifying factor 2 |
| chr12_-_34621359 | 0.27 |
|
|
|
| chr7_+_9628564 | 0.27 |
ENSDART00000172813
ENSDART00000128376 |
lins1
|
lines homolog 1 |
| chr11_-_6051096 | 0.27 |
ENSDART00000147761
|
vsg1
|
vessel-specific 1 |
| chr13_-_40027211 | 0.27 |
|
|
|
| chr6_+_23611854 | 0.27 |
ENSDART00000167795
|
pik3r6a
|
phosphoinositide-3-kinase, regulatory subunit 6a |
| chr11_-_6442836 | 0.26 |
ENSDART00000004483
|
zgc:162969
|
zgc:162969 |
| chr8_+_25015325 | 0.26 |
ENSDART00000140617
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr6_+_40924749 | 0.26 |
ENSDART00000133599
|
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr5_-_12706441 | 0.25 |
ENSDART00000051666
|
ppm1f
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr23_-_46268313 | 0.25 |
ENSDART00000170417
ENSDART00000168352 |
CABZ01102528.1
|
ENSDARG00000098990 |
| chr19_-_13946434 | 0.25 |
ENSDART00000166895
|
ctgfb
|
connective tissue growth factor b |
| chr21_-_40653250 | 0.25 |
ENSDART00000162623
|
arxb
|
aristaless related homeobox b |
| chr19_-_11188322 | 0.25 |
ENSDART00000059102
|
arhgef1a
|
Rho guanine nucleotide exchange factor (GEF) 1a |
| chr13_+_4096622 | 0.25 |
ENSDART00000058242
ENSDART00000143456 |
mea1
|
male-enhanced antigen 1 |
| chr10_-_26782374 | 0.24 |
ENSDART00000162710
|
fgf13b
|
fibroblast growth factor 13b |
| chr10_+_39211384 | 0.24 |
|
|
|
| chr21_+_26684728 | 0.24 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr22_-_2828572 | 0.24 |
|
|
|
| chr4_+_11385828 | 0.23 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.6 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.4 | 1.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.4 | 1.4 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.3 | 0.7 | GO:0030431 | sleep(GO:0030431) |
| 0.3 | 1.1 | GO:0040014 | regulation of multicellular organism growth(GO:0040014) |
| 0.2 | 0.7 | GO:0050886 | phasic smooth muscle contraction(GO:0014821) vascular smooth muscle contraction(GO:0014829) endocrine process(GO:0050886) |
| 0.2 | 0.7 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.2 | 2.0 | GO:0003315 | heart rudiment formation(GO:0003315) |
| 0.2 | 1.4 | GO:0021627 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.2 | 1.2 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.2 | 0.6 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.2 | 1.4 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.2 | 0.7 | GO:0021512 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.2 | 0.7 | GO:0022602 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) inflammatory response to wounding(GO:0090594) |
| 0.1 | 0.6 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.1 | 0.4 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 1.1 | GO:0007622 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.1 | 0.9 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 0.9 | GO:2000637 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.1 | 1.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 1.6 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.1 | 1.4 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.6 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 1.1 | GO:1904263 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 0.3 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.1 | 1.7 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.1 | 1.0 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.1 | 0.5 | GO:0033262 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.1 | 0.5 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 0.7 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.4 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.9 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 0.6 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 0.9 | GO:1900052 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.1 | 0.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 1.2 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 0.7 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.1 | 1.0 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 0.8 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 0.2 | GO:2000623 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.4 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 1.1 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.1 | 0.3 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 1.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 2.2 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.5 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.9 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.4 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.2 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.8 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 0.8 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.4 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.7 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.6 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.2 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.7 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.5 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.9 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 0.5 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.2 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0099590 | postsynaptic neurotransmitter receptor internalization(GO:0098884) neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.2 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.9 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.3 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.8 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 0.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.6 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.0 | 0.4 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.4 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.2 | 0.5 | GO:0071914 | prominosome(GO:0071914) |
| 0.2 | 0.7 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.1 | 0.9 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.5 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.5 | GO:0031431 | Dbf4-dependent protein kinase complex(GO:0031431) |
| 0.1 | 1.1 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 0.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.5 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.5 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.1 | 0.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.5 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 1.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.9 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.6 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.1 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.3 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.6 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 3.0 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 0.7 | GO:0022626 | cytosolic large ribosomal subunit(GO:0022625) cytosolic ribosome(GO:0022626) |
| 0.0 | 0.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0032587 | ruffle membrane(GO:0032587) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 0.5 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.1 | 0.6 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.1 | 1.1 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 1.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.9 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.7 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.6 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.1 | 0.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.5 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.3 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 1.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 0.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 3.9 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.1 | 0.5 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.1 | 1.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.1 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 1.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.0 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.4 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.4 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.8 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.0 | 0.6 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 2.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 2.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.0 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 0.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 2.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.9 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 2.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 1.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.9 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 0.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.0 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 0.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.6 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 1.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 2.2 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |