Project
DANIO-CODE
Navigation
Downloads
Results for lmx1ba+lmx1bb
Z-value: 0.49
Transcription factors associated with lmx1ba+lmx1bb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lmx1bb
|
ENSDARG00000068365 | LIM homeobox transcription factor 1, beta b |
|
lmx1ba
|
ENSDARG00000104815 | LIM homeobox transcription factor 1, beta a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lmx1bb | dr10_dc_chr8_-_33372638_33372708 | -0.26 | 3.2e-01 | Click! |
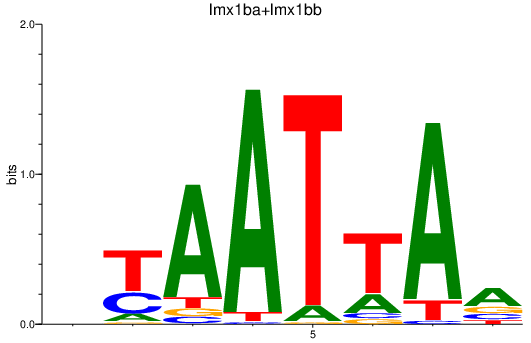
Activity profile of lmx1ba+lmx1bb motif
Sorted Z-values of lmx1ba+lmx1bb motif
Network of associatons between targets according to the STRING database.
First level regulatory network of lmx1ba+lmx1bb
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_34788205 | 1.27 |
ENSDART00000124800
|
fam212aa
|
family with sequence similarity 212, member Aa |
| chr2_+_33343287 | 0.89 |
ENSDART00000056655
|
klf17
|
Kruppel-like factor 17 |
| chr14_+_23420053 | 0.72 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr16_+_24046472 | 0.58 |
|
|
|
| chr14_+_23419894 | 0.53 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr23_-_31986679 | 0.51 |
ENSDART00000085054
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr23_-_31986571 | 0.46 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr7_+_19300351 | 0.43 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr2_+_10192670 | 0.42 |
ENSDART00000011906
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr14_+_23419864 | 0.40 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr18_+_19467527 | 0.39 |
ENSDART00000079695
|
zwilch
|
zwilch kinetochore protein |
| chr23_-_31986482 | 0.39 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr8_-_44247277 | 0.38 |
|
|
|
| chr8_-_23591082 | 0.36 |
ENSDART00000025024
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr21_+_26684617 | 0.34 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr8_+_45326435 | 0.33 |
ENSDART00000134161
|
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr10_+_6925975 | 0.33 |
|
|
|
| chr20_+_46588401 | 0.32 |
ENSDART00000060695
|
zc3h14
|
zinc finger CCCH-type containing 14 |
| KN150200v1_+_7782 | 0.32 |
|
|
|
| chr23_-_35691369 | 0.31 |
ENSDART00000142369
|
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr17_+_16082472 | 0.29 |
ENSDART00000133154
|
znf395a
|
zinc finger protein 395a |
| chr6_+_40925259 | 0.29 |
ENSDART00000002728
ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr22_-_2828572 | 0.28 |
|
|
|
| chr14_+_34150130 | 0.28 |
ENSDART00000132193
ENSDART00000141058 |
wnt8a
BX927327.1
|
wingless-type MMTV integration site family, member 8a ENSDARG00000105311 |
| chr20_-_34126039 | 0.27 |
ENSDART00000033817
|
scyl3
|
SCY1-like, kinase-like 3 |
| chr1_+_25750825 | 0.27 |
ENSDART00000046376
|
g3bp2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr3_-_13771322 | 0.25 |
ENSDART00000159177
ENSDART00000165174 |
si:dkey-61n16.5
|
si:dkey-61n16.5 |
| chr8_+_28362546 | 0.25 |
ENSDART00000062682
|
adipor1b
|
adiponectin receptor 1b |
| chr19_+_770488 | 0.24 |
ENSDART00000062518
|
gstr
|
glutathione S-transferase rho |
| chr5_-_56539892 | 0.24 |
|
|
|
| chr8_+_45326523 | 0.24 |
ENSDART00000145011
|
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr5_-_18459312 | 0.24 |
ENSDART00000145210
|
ankle2
|
ankyrin repeat and LEM domain containing 2 |
| chr11_+_34909393 | 0.23 |
ENSDART00000110839
|
mon1a
|
MON1 secretory trafficking family member A |
| chr1_+_21244242 | 0.22 |
ENSDART00000087729
|
kdm4c
|
lysine (K)-specific demethylase 4C |
| chr15_-_16241500 | 0.22 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr11_-_34258956 | 0.22 |
ENSDART00000114004
|
pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr7_-_26332194 | 0.21 |
ENSDART00000099003
|
plscr3b
|
phospholipid scramblase 3b |
| chr6_-_8156520 | 0.21 |
ENSDART00000081561
|
ilf3a
|
interleukin enhancer binding factor 3a |
| chr6_+_40924749 | 0.21 |
ENSDART00000133599
|
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr19_+_31297635 | 0.21 |
ENSDART00000139599
|
yars
|
tyrosyl-tRNA synthetase |
| chr8_-_23591293 | 0.21 |
ENSDART00000025024
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr10_-_32550351 | 0.20 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| chr19_+_42863138 | 0.20 |
ENSDART00000076938
|
pogza
|
pogo transposable element derived with ZNF domain a |
| chr16_-_29452509 | 0.20 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr5_-_13890202 | 0.19 |
ENSDART00000026120
|
ap3m2
|
adaptor-related protein complex 3, mu 2 subunit |
| chr7_-_15008685 | 0.19 |
ENSDART00000173048
|
ENSDARG00000023868
|
ENSDARG00000023868 |
| chr20_-_33272014 | 0.19 |
ENSDART00000178752
ENSDART00000047834 |
nbas
|
neuroblastoma amplified sequence |
| chr11_-_17620732 | 0.19 |
ENSDART00000154627
|
eogt
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr15_-_36490729 | 0.17 |
|
|
|
| chr7_-_15008648 | 0.17 |
ENSDART00000173048
|
ENSDARG00000023868
|
ENSDARG00000023868 |
| chr21_-_27584637 | 0.16 |
|
|
|
| chr10_-_8238422 | 0.16 |
ENSDART00000129467
|
dhx29
|
DEAH (Asp-Glu-Ala-His) box polypeptide 29 |
| chr13_+_27102308 | 0.16 |
ENSDART00000145901
|
rin2
|
Ras and Rab interactor 2 |
| chr9_-_52950815 | 0.16 |
ENSDART00000161667
|
smarcal1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr21_+_26684728 | 0.15 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr8_-_5834829 | 0.15 |
ENSDART00000179217
|
ENSDARG00000106522
|
ENSDARG00000106522 |
| chr7_-_26332236 | 0.14 |
ENSDART00000099003
|
plscr3b
|
phospholipid scramblase 3b |
| chr21_-_44707326 | 0.14 |
ENSDART00000013814
|
ammecr1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 homolog (human) |
| chr3_-_36080647 | 0.14 |
ENSDART00000025326
|
csnk1da
|
casein kinase 1, delta a |
| chr10_-_43070198 | 0.13 |
ENSDART00000099270
|
CU326366.2
|
ENSDARG00000068593 |
| chr23_+_26300882 | 0.13 |
|
|
|
| chr14_-_21717949 | 0.13 |
ENSDART00000137795
|
ssrp1a
|
structure specific recognition protein 1a |
| chr11_-_6442836 | 0.13 |
ENSDART00000004483
|
zgc:162969
|
zgc:162969 |
| chr16_-_29451902 | 0.13 |
|
|
|
| chr16_+_19728218 | 0.12 |
|
|
|
| chr14_+_20045365 | 0.12 |
ENSDART00000167637
|
aff2
|
AF4/FMR2 family, member 2 |
| chr10_+_17755665 | 0.12 |
ENSDART00000170453
|
slc20a1b
|
solute carrier family 20 (phosphate transporter), member 1b |
| chr8_+_30103357 | 0.12 |
ENSDART00000133717
|
fancc
|
Fanconi anemia, complementation group C |
| chr15_-_23587330 | 0.11 |
ENSDART00000167246
|
hmbsb
|
hydroxymethylbilane synthase, b |
| chr15_-_16241412 | 0.11 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr3_-_16569378 | 0.11 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr21_-_11877044 | 0.11 |
ENSDART00000145194
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr5_+_65754237 | 0.11 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr7_+_71757218 | 0.11 |
ENSDART00000012918
|
psmd9
|
proteasome 26S subunit, non-ATPase 9 |
| chr10_-_15382145 | 0.11 |
ENSDART00000133919
|
pum3
|
pumilio RNA-binding family member 3 |
| chr13_-_50032548 | 0.11 |
ENSDART00000038120
|
cacul1
|
CDK2 associated cullin domain 1 |
| chr5_-_68848113 | 0.10 |
ENSDART00000097249
|
aldh2.2
|
aldehyde dehydrogenase 2 family (mitochondrial), tandem duplicate 2 |
| chr7_+_8492338 | 0.10 |
|
|
|
| chr15_-_40391882 | 0.09 |
|
|
|
| chr23_-_29627060 | 0.09 |
ENSDART00000166554
|
ube4b
|
ubiquitination factor E4B, UFD2 homolog (S. cerevisiae) |
| chr13_+_27102377 | 0.09 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr5_-_50346132 | 0.09 |
ENSDART00000097460
|
hmgcra
|
3-hydroxy-3-methylglutaryl-CoA reductase a |
| chr7_+_7463292 | 0.09 |
ENSDART00000091099
|
ino80b
|
INO80 complex subunit B |
| chr16_-_21238288 | 0.08 |
ENSDART00000139737
|
cbx3b
|
chromobox homolog 3b |
| chr9_-_12623451 | 0.08 |
ENSDART00000142024
|
tra2b
|
transformer 2 beta homolog |
| chr23_-_25853364 | 0.08 |
ENSDART00000110670
|
ENSDARG00000078574
|
ENSDARG00000078574 |
| chr11_-_34909095 | 0.08 |
ENSDART00000017393
|
traip
|
TRAF-interacting protein |
| chr13_-_36015558 | 0.08 |
ENSDART00000133072
|
med6
|
mediator complex subunit 6 |
| chr18_-_43890836 | 0.08 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr18_-_40718244 | 0.08 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr8_+_25015374 | 0.08 |
ENSDART00000140617
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr17_-_2513630 | 0.08 |
ENSDART00000135374
|
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr1_+_43471754 | 0.07 |
ENSDART00000166324
|
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr23_-_25853220 | 0.07 |
ENSDART00000110670
|
ENSDARG00000078574
|
ENSDARG00000078574 |
| chr15_-_5912105 | 0.07 |
|
|
|
| chr11_+_17849528 | 0.07 |
ENSDART00000020283
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr14_-_6831373 | 0.07 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr14_-_6831308 | 0.07 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr1_+_43471683 | 0.07 |
ENSDART00000166324
|
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr11_+_17849608 | 0.06 |
ENSDART00000020283
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr5_+_20640468 | 0.06 |
ENSDART00000028087
|
cds2
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 |
| chr22_+_1153361 | 0.06 |
ENSDART00000159761
|
irf6
|
interferon regulatory factor 6 |
| chr12_-_45496246 | 0.06 |
ENSDART00000176229
ENSDART00000175457 |
FO818685.1
|
ENSDARG00000107895 |
| chr24_+_8702288 | 0.05 |
ENSDART00000114810
|
sycp2l
|
synaptonemal complex protein 2-like |
| chr12_+_22497249 | 0.05 |
ENSDART00000178120
|
CR848021.1
|
ENSDARG00000107088 |
| chr18_+_20571400 | 0.05 |
ENSDART00000151990
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr3_+_17387551 | 0.05 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr24_-_16922912 | 0.05 |
ENSDART00000111237
|
armc3
|
armadillo repeat containing 3 |
| chr24_+_28304003 | 0.05 |
ENSDART00000018095
|
sh3glb1a
|
SH3-domain GRB2-like endophilin B1a |
| chr11_-_30364626 | 0.05 |
ENSDART00000089803
ENSDART00000046800 ENSDART00000172244 |
slc8a1a
|
solute carrier family 8 (sodium/calcium exchanger), member 1a |
| chr17_+_23278879 | 0.05 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr20_+_47588954 | 0.05 |
ENSDART00000021341
|
kif3ca
|
kinesin family member 3Ca |
| chr11_-_37730181 | 0.05 |
ENSDART00000102870
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr3_+_22312446 | 0.04 |
ENSDART00000156450
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chr12_-_26062065 | 0.04 |
ENSDART00000076051
|
opn4b
|
opsin 4b |
| chr6_-_32001100 | 0.04 |
ENSDART00000132280
|
ror1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr22_-_17627831 | 0.04 |
ENSDART00000138483
|
si:ch73-243b8.4
|
si:ch73-243b8.4 |
| chr16_+_25202272 | 0.04 |
ENSDART00000163244
|
si:ch211-261d7.6
|
si:ch211-261d7.6 |
| chr22_+_1153570 | 0.04 |
ENSDART00000157442
|
irf6
|
interferon regulatory factor 6 |
| chr23_+_45486958 | 0.03 |
ENSDART00000111126
|
psip1b
|
PC4 and SFRS1 interacting protein 1b |
| chr20_-_23527004 | 0.03 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr16_-_31494275 | 0.03 |
ENSDART00000056551
|
csnk2a1
|
casein kinase 2, alpha 1 polypeptide |
| chr17_+_10437343 | 0.03 |
ENSDART00000167188
|
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr10_-_34971926 | 0.03 |
ENSDART00000141201
|
ccna1
|
cyclin A1 |
| chr13_+_33173790 | 0.03 |
ENSDART00000145295
|
dcdc2b
|
doublecortin domain containing 2B |
| chr14_-_6831278 | 0.03 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr4_+_13587809 | 0.03 |
ENSDART00000138201
|
tnpo3
|
transportin 3 |
| chr6_+_13833991 | 0.03 |
|
|
|
| chr3_+_48811662 | 0.03 |
ENSDART00000023814
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr10_+_39211384 | 0.03 |
|
|
|
| chr14_-_33605295 | 0.02 |
ENSDART00000168546
|
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr4_+_9668755 | 0.02 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr1_+_35253821 | 0.02 |
ENSDART00000179634
|
zgc:152968
|
zgc:152968 |
| chr24_-_14447773 | 0.02 |
|
|
|
| chr15_-_5911817 | 0.02 |
|
|
|
| chr19_-_42759951 | 0.02 |
|
|
|
| chr13_-_40027211 | 0.02 |
|
|
|
| chr14_-_3836967 | 0.02 |
ENSDART00000055817
|
ENSDARG00000038270
|
ENSDARG00000038270 |
| chr5_-_28693064 | 0.02 |
|
|
|
| chr3_-_26112995 | 0.02 |
ENSDART00000103741
|
ppp4ca
|
protein phosphatase 4, catalytic subunit a |
| chr1_-_25750237 | 0.02 |
ENSDART00000125690
|
ints12
|
integrator complex subunit 12 |
| chr22_-_17627900 | 0.02 |
ENSDART00000138483
|
si:ch73-243b8.4
|
si:ch73-243b8.4 |
| chr7_-_26331734 | 0.02 |
|
|
|
| chr21_-_40653250 | 0.02 |
ENSDART00000162623
|
arxb
|
aristaless related homeobox b |
| chr19_-_1921502 | 0.02 |
ENSDART00000004585
|
clptm1l
|
CLPTM1-like |
| chr24_-_35673629 | 0.02 |
ENSDART00000058564
|
mcm4
|
minichromosome maintenance complex component 4 |
| chr20_-_9107294 | 0.01 |
ENSDART00000140792
|
OMA1
|
OMA1 zinc metallopeptidase |
| chr17_+_9152727 | 0.01 |
|
|
|
| chr16_-_42105636 | 0.01 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr25_-_3742327 | 0.01 |
ENSDART00000075663
|
cracr2b
|
calcium release activated channel regulator 2B |
| chr25_-_19388381 | 0.01 |
ENSDART00000154986
|
zgc:193812
|
zgc:193812 |
| chr10_-_25448712 | 0.01 |
ENSDART00000140023
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr9_+_36316158 | 0.01 |
ENSDART00000176763
|
lrp1bb
|
low density lipoprotein receptor-related protein 1Bb |
| chr8_+_1834454 | 0.00 |
ENSDART00000056208
|
snap29
|
synaptosomal-associated protein 29 |
| chr3_-_29932835 | 0.00 |
|
|
|
| chr8_-_11512545 | 0.00 |
ENSDART00000133932
|
si:ch211-248e11.2
|
si:ch211-248e11.2 |
| chr13_-_37340209 | 0.00 |
|
|
|
| chr7_+_23752492 | 0.00 |
ENSDART00000141165
|
tinf2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr17_-_31562669 | 0.00 |
ENSDART00000110167
|
rpap1
|
RNA polymerase II associated protein 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.2 | 0.6 | GO:0089709 | serine transport(GO:0032329) L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.1 | 0.4 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.1 | 0.3 | GO:0039015 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.1 | 0.3 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 0.2 | GO:2000622 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.5 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.1 | 1.4 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 0.1 | GO:0001659 | temperature homeostasis(GO:0001659) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 1.6 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 0.2 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.4 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.4 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.2 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.1 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.0 | 0.1 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.0 | 0.2 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0035493 | SNARE complex assembly(GO:0035493) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.0 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.0 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.6 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.2 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.2 | GO:0008665 | tRNA 2'-phosphotransferase activity(GO:0000215) 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.4 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.2 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.2 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.1 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 0.2 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |