Project
DANIO-CODE
Navigation
Downloads
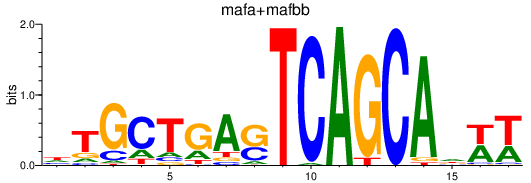
Results for mafa+mafbb
Z-value: 1.10
Transcription factors associated with mafa+mafbb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mafa
|
ENSDARG00000015890 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog a (paralog a) |
|
mafbb
|
ENSDARG00000070542 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Bb |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mafa | dr10_dc_chr18_+_29424354_29424406 | 0.88 | 6.8e-06 | Click! |
| mafbb | dr10_dc_chr11_-_25146564_25146598 | 0.68 | 3.8e-03 | Click! |
Activity profile of mafa+mafbb motif
Sorted Z-values of mafa+mafbb motif
Network of associatons between targets according to the STRING database.
First level regulatory network of mafa+mafbb
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_30699938 | 5.35 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr6_-_13654186 | 3.77 |
ENSDART00000150102
ENSDART00000041269 |
cryba2a
|
crystallin, beta A2a |
| chr24_-_12794564 | 3.65 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr5_-_66792947 | 3.21 |
ENSDART00000147009
|
si:dkey-251i10.2
|
si:dkey-251i10.2 |
| chr16_+_46327528 | 3.18 |
ENSDART00000040769
|
nr2f5
|
nuclear receptor subfamily 2, group F, member 5 |
| chr3_-_17881044 | 3.17 |
ENSDART00000013540
|
si:ch73-141c7.1
|
si:ch73-141c7.1 |
| chr15_-_29454583 | 3.16 |
ENSDART00000145976
|
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr13_+_2774422 | 3.10 |
ENSDART00000162362
|
wu:fj16a03
|
wu:fj16a03 |
| chr2_-_24898678 | 2.86 |
ENSDART00000145145
|
cnn2
|
calponin 2 |
| chr8_-_41194452 | 2.85 |
ENSDART00000165949
|
fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr11_-_5868257 | 2.63 |
ENSDART00000104360
|
gamt
|
guanidinoacetate N-methyltransferase |
| chr16_+_33702010 | 2.62 |
ENSDART00000143757
|
fhl3a
|
four and a half LIM domains 3a |
| chr14_-_45883839 | 2.57 |
|
|
|
| chr10_+_10843384 | 2.52 |
ENSDART00000130283
|
ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr13_-_33574216 | 2.49 |
ENSDART00000065435
|
cst3
|
cystatin C (amyloid angiopathy and cerebral hemorrhage) |
| chr20_-_14158114 | 2.49 |
ENSDART00000009549
|
rhag
|
Rh-associated glycoprotein |
| chr20_-_47123598 | 2.45 |
ENSDART00000100320
|
dnmt3aa
|
DNA (cytosine-5-)-methyltransferase 3 alpha a |
| chr22_-_6977038 | 2.37 |
ENSDART00000133143
ENSDART00000146813 |
gpd1b
|
glycerol-3-phosphate dehydrogenase 1b |
| chr20_-_29517770 | 2.34 |
ENSDART00000147464
|
ryr3
|
ryanodine receptor 3 |
| chr5_-_30015572 | 2.34 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr1_-_29706856 | 2.29 |
ENSDART00000174868
|
igf2bp2b
|
insulin-like growth factor 2 mRNA binding protein 2b |
| chr21_+_43204049 | 2.24 |
ENSDART00000151748
|
aff4
|
AF4/FMR2 family, member 4 |
| chr13_-_22877318 | 2.14 |
ENSDART00000057638
ENSDART00000171778 |
hk1
|
hexokinase 1 |
| chr12_+_16845837 | 2.12 |
ENSDART00000138174
|
slc16a12b
|
solute carrier family 16, member 12b |
| chr23_-_26150495 | 2.11 |
ENSDART00000126299
|
gdi1
|
GDP dissociation inhibitor 1 |
| chr21_-_28486800 | 2.08 |
ENSDART00000077910
|
epdl2
|
ependymin-like 2 |
| chr2_-_9950224 | 2.02 |
ENSDART00000146407
|
si:ch1073-170o4.1
|
si:ch1073-170o4.1 |
| chr9_-_30437160 | 1.95 |
ENSDART00000147241
|
BX936337.1
|
ENSDARG00000092870 |
| chr3_+_27684309 | 1.89 |
|
|
|
| chr23_-_31502149 | 1.86 |
ENSDART00000053545
|
zgc:153284
|
zgc:153284 |
| chr7_+_13753344 | 1.81 |
|
|
|
| chr19_+_32183937 | 1.77 |
ENSDART00000147474
|
stmn2b
|
stathmin 2b |
| chr9_-_12653481 | 1.73 |
ENSDART00000122558
|
tspearb
|
thrombospondin-type laminin G domain and EAR repeats b |
| chr2_+_2259793 | 1.69 |
ENSDART00000109073
|
dsg2l
|
desmoglein 2 like |
| chr24_-_12794672 | 1.61 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr7_+_39116005 | 1.54 |
ENSDART00000146702
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr10_+_31336284 | 1.54 |
ENSDART00000140988
|
tmem218
|
transmembrane protein 218 |
| chr14_+_4169846 | 1.51 |
ENSDART00000038301
|
gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr23_-_20837809 | 1.44 |
ENSDART00000089750
|
znf362b
|
zinc finger protein 362b |
| chr6_-_39311711 | 1.42 |
|
|
|
| chr1_-_11591843 | 1.37 |
ENSDART00000003825
|
cplx2l
|
complexin 2, like |
| chr14_+_34207218 | 1.36 |
ENSDART00000135608
|
gabrp
|
gamma-aminobutyric acid (GABA) A receptor, pi |
| chr11_+_666006 | 1.34 |
ENSDART00000130839
|
timp4.1
|
TIMP metallopeptidase inhibitor 4, tandem duplicate 1 |
| chr21_+_21337628 | 1.32 |
ENSDART00000136325
|
rtn2b
|
reticulon 2b |
| chr8_-_41194530 | 1.25 |
ENSDART00000173055
|
fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr7_-_18791134 | 1.24 |
ENSDART00000158796
|
fcer1g
|
Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide |
| chr8_+_28513670 | 1.22 |
ENSDART00000110291
|
srxn1
|
sulfiredoxin 1 homolog (S. cerevisiae) |
| chr7_+_42124857 | 1.21 |
ENSDART00000004120
|
adamts18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18 |
| chr3_+_25992836 | 1.17 |
ENSDART00000010477
|
hsp70.3
|
heat shock cognate 70-kd protein, tandem duplicate 3 |
| chr16_-_30606364 | 1.16 |
ENSDART00000147986
|
lmna
|
lamin A |
| KN150670v1_-_32201 | 1.15 |
|
|
|
| chr2_-_21960233 | 1.13 |
ENSDART00000027587
|
ralab
|
v-ral simian leukemia viral oncogene homolog Ab (ras related) |
| chr11_-_33697442 | 1.12 |
ENSDART00000087597
|
col6a2
|
collagen, type VI, alpha 2 |
| chr18_+_14627030 | 1.11 |
ENSDART00000080788
|
wfdc1
|
WAP four-disulfide core domain 1 |
| chr12_-_3903961 | 1.09 |
ENSDART00000134292
|
ENSDARG00000021154
|
ENSDARG00000021154 |
| chr7_-_25426346 | 1.09 |
ENSDART00000082620
|
dysf
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive) |
| chr14_+_34440699 | 1.09 |
ENSDART00000130469
|
sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr5_-_18548689 | 1.06 |
ENSDART00000137802
|
fam214b
|
family with sequence similarity 214, member B |
| chr5_-_37335445 | 1.05 |
ENSDART00000141233
|
scn4bb
|
sodium channel, voltage-gated, type IV, beta b |
| chr8_-_50299273 | 1.05 |
ENSDART00000023639
|
nkx2.7
|
NK2 transcription factor related 7 |
| chr1_-_11591684 | 1.02 |
ENSDART00000003825
|
cplx2l
|
complexin 2, like |
| chr5_-_64315770 | 0.99 |
ENSDART00000169287
|
zgc:110283
|
zgc:110283 |
| chr16_+_13964949 | 0.97 |
ENSDART00000143983
|
zgc:174888
|
zgc:174888 |
| chr23_+_7445760 | 0.97 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr11_-_39864306 | 0.96 |
ENSDART00000165781
|
fam83e
|
family with sequence similarity 83, member E |
| chr7_+_23604092 | 0.95 |
ENSDART00000101406
|
rab39bb
|
RAB39B, member RAS oncogene family b |
| chr20_+_18762031 | 0.95 |
ENSDART00000152136
ENSDART00000126959 |
tnfaip2a
|
tumor necrosis factor, alpha-induced protein 2a |
| chr3_-_28120668 | 0.94 |
|
|
|
| chr13_-_22901327 | 0.93 |
ENSDART00000056523
|
hkdc1
|
hexokinase domain containing 1 |
| chr10_+_26786051 | 0.90 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr12_-_28671833 | 0.88 |
|
|
|
| chr1_-_51862897 | 0.87 |
ENSDART00000136469
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr16_+_7745309 | 0.87 |
ENSDART00000081418
ENSDART00000149404 |
bves
|
blood vessel epicardial substance |
| chr3_+_27477793 | 0.86 |
|
|
|
| chr14_+_34440799 | 0.86 |
ENSDART00000173371
|
sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr12_+_7421078 | 0.86 |
ENSDART00000163114
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr15_-_2079036 | 0.80 |
ENSDART00000112946
|
dock10
|
dedicator of cytokinesis 10 |
| chr1_+_18610282 | 0.78 |
ENSDART00000129970
|
si:dkeyp-118a3.2
|
si:dkeyp-118a3.2 |
| chr11_-_39864543 | 0.78 |
ENSDART00000165781
|
fam83e
|
family with sequence similarity 83, member E |
| chr20_+_18841440 | 0.77 |
ENSDART00000174532
|
fam167ab
|
family with sequence similarity 167, member Ab |
| chr6_-_39311594 | 0.76 |
|
|
|
| chr17_-_50343403 | 0.75 |
ENSDART00000140121
|
si:ch211-235i11.5
|
si:ch211-235i11.5 |
| chr2_+_26161204 | 0.74 |
ENSDART00000078639
|
slc2a2
|
solute carrier family 2 (facilitated glucose transporter), member 2 |
| chr16_-_31476944 | 0.69 |
ENSDART00000167350
|
zgc:194210
|
zgc:194210 |
| chr24_+_21876490 | 0.68 |
ENSDART00000081234
|
ropn1l
|
rhophilin associated tail protein 1-like |
| chr6_-_40747134 | 0.65 |
ENSDART00000154916
|
p4htm
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr18_+_17548213 | 0.62 |
ENSDART00000144960
|
nup93
|
nucleoporin 93 |
| chr4_+_5308883 | 0.62 |
ENSDART00000150366
|
si:ch211-214j24.10
|
si:ch211-214j24.10 |
| chr24_-_42120772 | 0.60 |
ENSDART00000166413
ENSDART00000166414 |
ssr1
|
signal sequence receptor, alpha |
| chr2_+_38390887 | 0.59 |
ENSDART00000113111
|
psmb5
|
proteasome subunit beta 5 |
| chr19_-_9793444 | 0.58 |
ENSDART00000134816
|
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr10_+_877701 | 0.57 |
ENSDART00000176303
|
FO704646.1
|
ENSDARG00000107355 |
| chr10_+_13320935 | 0.57 |
ENSDART00000135082
|
tmem267
|
transmembrane protein 267 |
| chr18_-_16934961 | 0.57 |
ENSDART00000122102
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr21_+_19311031 | 0.57 |
ENSDART00000093155
|
hpse
|
heparanase |
| chr7_-_6280179 | 0.55 |
ENSDART00000173349
|
CU457819.3
|
Histone H3.2 |
| chr11_-_40415291 | 0.50 |
|
|
|
| chr11_+_23012136 | 0.48 |
ENSDART00000110152
|
csf1a
|
colony stimulating factor 1a (macrophage) |
| chr19_+_7535067 | 0.46 |
ENSDART00000147491
|
fam63a
|
family with sequence similarity 63, member A |
| chr18_+_12452 | 0.45 |
|
|
|
| chr17_-_14956738 | 0.43 |
|
|
|
| chr19_+_32184149 | 0.43 |
ENSDART00000078299
|
stmn2b
|
stathmin 2b |
| chr5_-_38022706 | 0.43 |
|
|
|
| chr4_+_23406078 | 0.41 |
ENSDART00000139543
|
mdm2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr9_-_12063411 | 0.39 |
ENSDART00000038651
|
znf804a
|
zinc finger protein 804A |
| chr11_+_44922098 | 0.37 |
ENSDART00000172999
|
mafgb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
| chr4_+_15607042 | 0.37 |
ENSDART00000101619
ENSDART00000021384 |
exoc4
|
exocyst complex component 4 |
| chr8_-_14014576 | 0.36 |
ENSDART00000135811
|
atp2b3a
|
ATPase, Ca++ transporting, plasma membrane 3a |
| chr19_+_7535152 | 0.35 |
ENSDART00000136502
|
fam63a
|
family with sequence similarity 63, member A |
| chr11_+_40280114 | 0.34 |
ENSDART00000175424
|
mmel1
|
membrane metallo-endopeptidase-like 1 |
| chr1_-_11591631 | 0.34 |
ENSDART00000142122
|
cplx2l
|
complexin 2, like |
| chr3_+_15245529 | 0.33 |
|
|
|
| chr17_+_33862300 | 0.33 |
|
|
|
| chr10_+_877636 | 0.33 |
ENSDART00000176303
|
FO704646.1
|
ENSDARG00000107355 |
| chr3_-_28120760 | 0.32 |
|
|
|
| chr1_-_6403371 | 0.31 |
|
|
|
| chr12_-_151380 | 0.31 |
|
|
|
| chr8_+_36888422 | 0.30 |
|
|
|
| chr22_-_38530581 | 0.30 |
ENSDART00000151785
|
klc4
|
kinesin light chain 4 |
| chr4_+_13451169 | 0.29 |
ENSDART00000172552
|
dyrk2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr17_-_31562669 | 0.28 |
ENSDART00000110167
|
rpap1
|
RNA polymerase II associated protein 1 |
| KN150670v1_-_31884 | 0.27 |
|
|
|
| chr8_+_5546852 | 0.27 |
|
|
|
| chr10_+_38764480 | 0.26 |
ENSDART00000172306
ENSDART00000163099 |
tmprss2
|
transmembrane protease, serine 2 |
| chr11_-_39864489 | 0.25 |
ENSDART00000165781
|
fam83e
|
family with sequence similarity 83, member E |
| KN149949v1_+_25867 | 0.25 |
|
|
|
| chr14_+_26424717 | 0.25 |
|
|
|
| chr20_-_24047119 | 0.24 |
ENSDART00000143005
|
mdn1
|
midasin AAA ATPase 1 |
| chr14_-_2284860 | 0.23 |
ENSDART00000124171
|
si:dkeyp-115d7.2
|
si:dkeyp-115d7.2 |
| KN149949v1_+_25953 | 0.23 |
|
|
|
| chr21_-_45647945 | 0.23 |
|
|
|
| chr12_-_30898555 | 0.23 |
|
|
|
| chr25_-_34540270 | 0.22 |
ENSDART00000156508
|
zgc:114046
|
zgc:114046 |
| chr24_+_3177066 | 0.22 |
|
|
|
| chr14_+_4169371 | 0.21 |
ENSDART00000136665
|
gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr8_-_11732343 | 0.21 |
ENSDART00000091684
|
anapc7
|
anaphase promoting complex subunit 7 |
| KN149797v1_-_70680 | 0.20 |
ENSDART00000170785
|
CABZ01113193.1
|
uncharacterized protein LOC795664 precursor |
| chr12_-_30898994 | 0.19 |
|
|
|
| chr23_-_3467855 | 0.17 |
|
|
|
| chr6_-_26905146 | 0.14 |
ENSDART00000011863
|
hdlbpa
|
high density lipoprotein binding protein a |
| chr1_-_51863187 | 0.14 |
ENSDART00000004233
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr18_+_10957421 | 0.13 |
ENSDART00000166561
|
ttc38
|
tetratricopeptide repeat domain 38 |
| chr13_-_25068717 | 0.13 |
ENSDART00000057605
|
adka
|
adenosine kinase a |
| chr15_+_29842298 | 0.13 |
|
|
|
| chr12_-_1018688 | 0.11 |
ENSDART00000054353
|
ENSDARG00000037357
|
ENSDARG00000037357 |
| chr11_+_42474134 | 0.11 |
ENSDART00000156080
|
tdrd3
|
tudor domain containing 3 |
| chr19_-_35841625 | 0.11 |
ENSDART00000169006
|
ak2
|
adenylate kinase 2 |
| chr11_-_40414664 | 0.10 |
ENSDART00000165163
|
si:ch211-222l21.1
|
si:ch211-222l21.1 |
| chr17_-_51173358 | 0.09 |
ENSDART00000169050
|
trappc12
|
trafficking protein particle complex 12 |
| chr24_+_14095643 | 0.09 |
ENSDART00000124740
|
ncoa2
|
nuclear receptor coactivator 2 |
| chr3_+_27188961 | 0.08 |
|
|
|
| chr5_-_30434967 | 0.08 |
ENSDART00000086534
|
cyb5d2
|
cytochrome b5 domain containing 2 |
| chr20_+_54544931 | 0.08 |
|
|
|
| chr21_-_5228529 | 0.07 |
ENSDART00000146061
|
psmd5
|
proteasome 26S subunit, non-ATPase 5 |
| chr21_-_43332911 | 0.06 |
ENSDART00000114955
|
sowahaa
|
sosondowah ankyrin repeat domain family member Aa |
| chr4_-_23405867 | 0.05 |
|
|
|
| chr12_+_33716314 | 0.04 |
ENSDART00000111486
|
mrpl43
|
mitochondrial ribosomal protein L43 |
| chr23_-_887934 | 0.03 |
|
|
|
| chr14_+_20927461 | 0.03 |
ENSDART00000160576
|
si:ch211-175m2.4
|
si:ch211-175m2.4 |
| chr17_-_31027949 | 0.03 |
ENSDART00000152016
|
eml1
|
echinoderm microtubule associated protein like 1 |
| chr7_+_49780512 | 0.03 |
ENSDART00000174308
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr12_-_3904030 | 0.02 |
ENSDART00000134292
|
ENSDARG00000021154
|
ENSDARG00000021154 |
| chr7_+_41141295 | 0.01 |
ENSDART00000083967
|
chpf2
|
chondroitin polymerizing factor 2 |
| chr6_-_33940469 | 0.00 |
ENSDART00000137268
|
akr1a1b
|
aldo-keto reductase family 1, member A1b (aldehyde reductase) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.3 | GO:0071549 | propionate metabolic process(GO:0019541) propionate catabolic process(GO:0019543) glycerol biosynthetic process from pyruvate(GO:0046327) cellular response to dexamethasone stimulus(GO:0071549) |
| 0.9 | 2.6 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.8 | 2.5 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.5 | 2.4 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.4 | 1.3 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.4 | 1.1 | GO:0002280 | monocyte activation involved in immune response(GO:0002280) negative regulation of phagocytosis(GO:0050765) |
| 0.3 | 1.7 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.3 | 2.1 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.3 | 6.9 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 1.0 | GO:0009886 | post-embryonic morphogenesis(GO:0009886) post-embryonic foregut morphogenesis(GO:0048618) |
| 0.2 | 2.9 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.2 | 0.9 | GO:0001919 | regulation of receptor recycling(GO:0001919) response to ischemia(GO:0002931) |
| 0.2 | 2.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.2 | 3.2 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.2 | 0.7 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.2 | 1.2 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.2 | 1.0 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.2 | 0.6 | GO:0051292 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.2 | 3.2 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.4 | GO:1904035 | epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) |
| 0.1 | 1.0 | GO:0003207 | cardiac chamber formation(GO:0003207) |
| 0.1 | 2.3 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.1 | 1.2 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 2.7 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 0.4 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 0.7 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 2.2 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 2.5 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.1 | 2.2 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 3.8 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 1.0 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 0.7 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 | 2.0 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 1.4 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 2.1 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 2.1 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.7 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.4 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.9 | GO:0007596 | blood coagulation(GO:0007596) hemostasis(GO:0007599) |
| 0.0 | 0.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.0 | 1.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.1 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.0 | 0.2 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 1.1 | GO:0048747 | muscle fiber development(GO:0048747) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.4 | 2.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.3 | 6.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.3 | 1.4 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.2 | 2.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.2 | 2.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 0.4 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 1.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 1.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 1.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 2.2 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 2.6 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 0.6 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.6 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 10.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.7 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 2.5 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 1.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.2 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.3 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.5 | 3.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.5 | 2.4 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.4 | 2.1 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.4 | 2.3 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.4 | 2.1 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.3 | 1.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 1.7 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.3 | 2.5 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.3 | 1.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 2.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.2 | 3.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 2.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 3.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 2.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.7 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 0.7 | GO:0015149 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.1 | 1.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 4.1 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 0.9 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 1.9 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 2.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 3.4 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 0.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 3.2 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 1.2 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.0 | 1.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 2.3 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.4 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 2.0 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.9 | GO:0008757 | S-adenosylmethionine-dependent methyltransferase activity(GO:0008757) |
| 0.0 | 1.0 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 2.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 7.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 5.1 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 0.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.3 | 2.5 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.2 | 5.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 0.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 2.8 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.2 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 2.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 0.6 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 2.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 2.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 3.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |