Project
DANIO-CODE
Navigation
Downloads
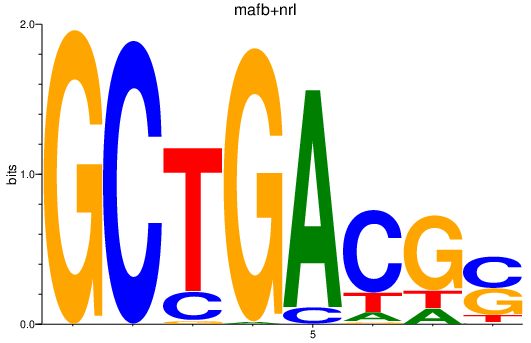
Results for mafb+nrl
Z-value: 1.59
Transcription factors associated with mafb+nrl
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mafb
|
ENSDARG00000076520 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog b (paralog b) |
|
nrl
|
ENSDARG00000100466 | neural retina leucine zipper |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nrl | dr10_dc_chr23_-_45707212_45707216 | 0.84 | 4.6e-05 | Click! |
Activity profile of mafb+nrl motif
Sorted Z-values of mafb+nrl motif
Network of associatons between targets according to the STRING database.
First level regulatory network of mafb+nrl
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_70948223 | 3.53 |
ENSDART00000013404
|
ak1
|
adenylate kinase 1 |
| chr3_-_54893038 | 3.43 |
ENSDART00000155871
ENSDART00000109016 |
hbae3
|
hemoglobin alpha embryonic-3 |
| chr22_-_613948 | 3.22 |
ENSDART00000148692
|
apobec2a
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2a |
| chr1_+_43738350 | 2.94 |
ENSDART00000127087
ENSDART00000100309 |
crybb1l2
|
crystallin, beta B1, like 2 |
| chr1_-_58580939 | 2.84 |
ENSDART00000158011
|
col5a3b
|
collagen, type V, alpha 3b |
| chr19_-_5429428 | 2.82 |
ENSDART00000066620
ENSDART00000151398 |
krtt1c19e
|
keratin type 1 c19e |
| chr20_+_12559243 | 2.78 |
ENSDART00000172370
|
si:dkey-269i1.4
|
si:dkey-269i1.4 |
| chr21_+_21337628 | 2.67 |
ENSDART00000136325
|
rtn2b
|
reticulon 2b |
| chr3_-_32686790 | 2.43 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr3_-_60953138 | 2.30 |
ENSDART00000028219
|
pvalb4
|
parvalbumin 4 |
| chr16_+_5470544 | 2.28 |
|
|
|
| chr21_+_40082890 | 2.27 |
ENSDART00000100166
|
serpinf1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| KN150074v1_-_1031 | 2.25 |
|
|
|
| chr21_-_99353 | 2.24 |
ENSDART00000040422
|
bhmt
|
betaine-homocysteine methyltransferase |
| chr24_+_38371710 | 2.23 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr7_+_58718614 | 2.22 |
ENSDART00000157873
|
hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr9_-_7560915 | 2.21 |
ENSDART00000081553
|
desma
|
desmin a |
| chr7_-_24428781 | 2.19 |
ENSDART00000052802
|
calb2b
|
calbindin 2b |
| chr7_+_38895198 | 2.19 |
ENSDART00000052318
|
mdka
|
midkine a |
| chr23_-_26150495 | 2.14 |
ENSDART00000126299
|
gdi1
|
GDP dissociation inhibitor 1 |
| chr19_-_5338129 | 2.07 |
ENSDART00000081951
|
stx1b
|
syntaxin 1B |
| chr16_-_39951493 | 1.98 |
|
|
|
| chr7_-_25816549 | 1.95 |
ENSDART00000052989
ENSDART00000047951 |
ache
|
acetylcholinesterase |
| chr11_+_14142126 | 1.88 |
ENSDART00000102520
|
palm1a
|
paralemmin 1a |
| chr7_-_25816655 | 1.86 |
ENSDART00000052989
ENSDART00000047951 |
ache
|
acetylcholinesterase |
| chr21_+_41684338 | 1.77 |
ENSDART00000169511
|
ppp2r2bb
|
protein phosphatase 2, regulatory subunit B, beta b |
| chr6_-_60152693 | 1.76 |
ENSDART00000083453
|
slc32a1
|
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr10_+_38831967 | 1.76 |
ENSDART00000125045
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr15_-_31687 | 1.73 |
|
|
|
| chr6_-_60152594 | 1.73 |
ENSDART00000083453
|
slc32a1
|
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr24_-_26138298 | 1.68 |
ENSDART00000128618
|
and1
|
actinodin1 |
| chr7_-_35438739 | 1.66 |
ENSDART00000043857
|
irx5a
|
iroquois homeobox 5a |
| chr24_-_12794672 | 1.65 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr9_-_42894582 | 1.64 |
ENSDART00000144744
|
col5a2a
|
collagen, type V, alpha 2a |
| chr6_+_55022668 | 1.63 |
ENSDART00000158845
|
mybphb
|
myosin binding protein Hb |
| chr3_-_28120668 | 1.63 |
|
|
|
| chr20_+_45531190 | 1.62 |
|
|
|
| chr22_+_26543146 | 1.61 |
|
|
|
| chr22_-_120677 | 1.58 |
|
|
|
| chr24_-_41448993 | 1.58 |
ENSDART00000041349
|
crygn2
|
crystallin, gamma N2 |
| chr6_-_39315024 | 1.57 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr7_+_16583234 | 1.57 |
ENSDART00000173580
|
nav2a
|
neuron navigator 2a |
| chr14_+_33117964 | 1.56 |
ENSDART00000075278
|
atp1b4
|
ATPase, Na+/K+ transporting, beta 4 polypeptide |
| chr19_-_5416317 | 1.51 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr15_-_31871 | 1.48 |
|
|
|
| chr8_-_38126693 | 1.46 |
ENSDART00000112331
|
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr7_+_50491026 | 1.45 |
ENSDART00000165037
|
si:ch73-380l10.2
|
si:ch73-380l10.2 |
| chr9_-_22371512 | 1.44 |
ENSDART00000101809
|
crygm2d6
|
crystallin, gamma M2d6 |
| chr9_-_22507592 | 1.44 |
ENSDART00000132029
|
crygm2d2
|
crystallin, gamma M2d2 |
| chr19_+_48499602 | 1.44 |
|
|
|
| chr10_-_26782374 | 1.43 |
ENSDART00000162710
|
fgf13b
|
fibroblast growth factor 13b |
| chr24_-_12794564 | 1.42 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr12_-_7790348 | 1.41 |
ENSDART00000148673
|
ank3b
|
ankyrin 3b |
| chr17_+_29328653 | 1.38 |
ENSDART00000086164
|
kctd3
|
potassium channel tetramerization domain containing 3 |
| chr20_-_21031504 | 1.38 |
ENSDART00000152726
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr21_-_34623741 | 1.37 |
ENSDART00000023038
|
dacha
|
dachshund a |
| chr15_-_4322875 | 1.36 |
ENSDART00000173311
|
si:ch211-117a13.2
|
si:ch211-117a13.2 |
| chr20_-_5009037 | 1.35 |
ENSDART00000164274
|
arid1b
|
AT rich interactive domain 1B (SWI1-like) |
| chr2_+_393439 | 1.34 |
ENSDART00000122138
|
mylk4a
|
myosin light chain kinase family, member 4a |
| chr12_+_7421078 | 1.34 |
ENSDART00000163114
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr19_+_33235375 | 1.34 |
ENSDART00000052096
|
hrsp12
|
heat-responsive protein 12 |
| chr23_-_3466041 | 1.33 |
ENSDART00000002309
|
mafba
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ba |
| chr25_+_4918339 | 1.33 |
ENSDART00000153980
|
parvb
|
parvin, beta |
| chr17_-_45383925 | 1.32 |
|
|
|
| chr12_+_27032862 | 1.31 |
|
|
|
| chr22_+_12327849 | 1.29 |
ENSDART00000178678
|
FO704572.1
|
ENSDARG00000106890 |
| chr6_-_39311711 | 1.28 |
|
|
|
| chr9_+_31079790 | 1.28 |
ENSDART00000101070
|
dachd
|
dachshund d |
| chr10_+_38831925 | 1.27 |
ENSDART00000125045
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr12_-_308929 | 1.27 |
ENSDART00000066579
ENSDART00000171396 |
pts
|
6-pyruvoyltetrahydropterin synthase |
| chr5_-_20527105 | 1.27 |
ENSDART00000133461
|
camk2b1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 1 |
| chr14_-_41311458 | 1.27 |
ENSDART00000163039
|
fgfrl1b
|
fibroblast growth factor receptor-like 1b |
| chr22_-_10430130 | 1.26 |
ENSDART00000064801
|
ogn
|
osteoglycin |
| chr9_+_39217373 | 1.26 |
ENSDART00000004742
|
cps1
|
carbamoyl-phosphate synthase 1, mitochondrial |
| chr17_+_44666606 | 1.25 |
ENSDART00000155096
|
tmem63c
|
transmembrane protein 63C |
| chr12_-_37921731 | 1.25 |
|
|
|
| chr9_+_25964943 | 1.23 |
ENSDART00000147229
ENSDART00000127834 |
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr13_-_49826330 | 1.23 |
ENSDART00000099475
|
nid1a
|
nidogen 1a |
| chr13_-_869704 | 1.23 |
|
|
|
| chr16_+_46145286 | 1.22 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr16_+_24069711 | 1.20 |
ENSDART00000142869
|
apoc2
|
apolipoprotein C-II |
| chr6_+_58549236 | 1.19 |
ENSDART00000157018
|
stmn3
|
stathmin-like 3 |
| chr16_-_42990753 | 1.19 |
ENSDART00000149317
|
hfe2
|
hemochromatosis type 2 |
| chr1_-_24747855 | 1.19 |
ENSDART00000111686
|
fhdc1
|
FH2 domain containing 1 |
| chr21_-_43020159 | 1.18 |
ENSDART00000065097
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr21_-_1600125 | 1.18 |
ENSDART00000066621
|
vmo1b
|
vitelline membrane outer layer 1 homolog b |
| chr4_+_26507297 | 1.16 |
ENSDART00000160652
|
iqsec3a
|
IQ motif and Sec7 domain 3a |
| KN149978v1_-_10669 | 1.16 |
ENSDART00000175723
|
CABZ01068178.1
|
ENSDARG00000105971 |
| chr5_-_63487161 | 1.15 |
ENSDART00000048395
|
cmlc1
|
cardiac myosin light chain-1 |
| chr8_+_50994889 | 1.15 |
ENSDART00000142061
|
si:dkey-32e23.4
|
si:dkey-32e23.4 |
| chr15_+_24277664 | 1.15 |
ENSDART00000155502
|
sez6b
|
seizure related 6 homolog b |
| chr4_+_3970874 | 1.14 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr25_-_34748472 | 1.14 |
|
|
|
| chr17_-_50140718 | 1.14 |
ENSDART00000058707
|
jdp2a
|
Jun dimerization protein 2a |
| chr2_+_31974269 | 1.13 |
ENSDART00000012413
|
ankhb
|
ANKH inorganic pyrophosphate transport regulator b |
| chr15_-_41288480 | 1.13 |
ENSDART00000155359
|
smco4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr24_-_475010 | 1.13 |
ENSDART00000108762
|
vopp1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr13_-_26907910 | 1.12 |
ENSDART00000146712
|
ccdc85a
|
coiled-coil domain containing 85A |
| chr13_-_25153833 | 1.12 |
ENSDART00000039828
ENSDART00000110304 |
vcla
|
vinculin a |
| chr9_-_29056763 | 1.12 |
ENSDART00000060321
|
ENSDARG00000056704
|
ENSDARG00000056704 |
| chr9_+_49961721 | 1.12 |
|
|
|
| chr10_+_1384178 | 1.12 |
ENSDART00000150096
|
gdnfa
|
glial cell derived neurotrophic factor a |
| chr8_-_14014576 | 1.11 |
ENSDART00000135811
|
atp2b3a
|
ATPase, Ca++ transporting, plasma membrane 3a |
| chr6_-_39312306 | 1.11 |
|
|
|
| chr9_+_21908880 | 1.10 |
|
|
|
| chr23_+_25782195 | 1.09 |
ENSDART00000060059
|
rbms2b
|
RNA binding motif, single stranded interacting protein 2b |
| chr8_-_24991984 | 1.08 |
ENSDART00000078795
|
ahcyl1
|
adenosylhomocysteinase-like 1 |
| chr18_+_45893199 | 1.08 |
ENSDART00000158246
|
dvl3b
|
dishevelled segment polarity protein 3b |
| chr16_-_48672056 | 1.07 |
|
|
|
| chr23_+_23559246 | 1.07 |
ENSDART00000172214
|
agrn
|
agrin |
| chr2_+_3864843 | 1.06 |
ENSDART00000108964
ENSDART00000128514 |
egfra
|
epidermal growth factor receptor a (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) |
| chr18_+_20504980 | 1.06 |
ENSDART00000060295
|
rapsn
|
receptor-associated protein of the synapse, 43kD |
| chr19_+_1797470 | 1.06 |
|
|
|
| chr2_-_44402486 | 1.05 |
ENSDART00000011188
ENSDART00000093298 |
atp1a2a
|
ATPase, Na+/K+ transporting, alpha 2a polypeptide |
| chr4_+_377829 | 1.05 |
ENSDART00000030215
|
rpl18a
|
ribosomal protein L18a |
| chr25_+_16505438 | 1.05 |
ENSDART00000008986
|
atp6v1e1a
|
ATPase, H+ transporting, lysosomal, V1 subunit E1a |
| chr7_+_10792973 | 1.05 |
|
|
|
| chr8_-_50899451 | 1.05 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long-chain family member 2 |
| chr16_+_5625301 | 1.04 |
|
|
|
| chr5_+_296856 | 1.04 |
|
|
|
| chr5_-_12025080 | 1.03 |
ENSDART00000164190
|
ksr2
|
kinase suppressor of ras 2 |
| chr1_-_43733382 | 1.02 |
ENSDART00000023642
|
cryba1l2
|
crystallin, beta A1, like 2 |
| chr21_-_5852663 | 1.01 |
ENSDART00000130521
|
CACFD1
|
calcium channel flower domain containing 1 |
| chr4_-_25847471 | 1.01 |
ENSDART00000142491
|
ndufa12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| KN150108v1_+_18276 | 1.00 |
|
|
|
| chr9_-_44493074 | 1.00 |
ENSDART00000167685
|
neurod1
|
neuronal differentiation 1 |
| chr9_+_54692834 | 1.00 |
|
|
|
| chr5_-_1698166 | 0.99 |
ENSDART00000073462
|
rplp0
|
ribosomal protein, large, P0 |
| chr14_-_16816075 | 0.99 |
ENSDART00000060479
|
smtnl1
|
smoothelin-like 1 |
| chr4_-_22130388 | 0.99 |
ENSDART00000019748
|
lin7a
|
lin-7 homolog A (C. elegans) |
| KN149760v1_-_29067 | 0.99 |
|
|
|
| chr4_+_19545842 | 0.99 |
ENSDART00000140028
|
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr6_-_60088120 | 0.99 |
|
|
|
| chr24_-_4941954 | 0.98 |
ENSDART00000127597
|
zic1
|
zic family member 1 (odd-paired homolog, Drosophila) |
| chr4_+_119964 | 0.98 |
ENSDART00000177177
ENSDART00000159996 |
tbk1
|
TANK-binding kinase 1 |
| chr25_+_30838768 | 0.98 |
|
|
|
| chr19_+_9085377 | 0.98 |
ENSDART00000091443
|
ENSDARG00000062900
|
ENSDARG00000062900 |
| chr13_-_24248675 | 0.97 |
ENSDART00000046360
|
rhoua
|
ras homolog family member Ua |
| chr25_+_36872560 | 0.97 |
ENSDART00000163178
|
slc10a3
|
solute carrier family 10, member 3 |
| chr13_-_18812889 | 0.97 |
|
|
|
| chr7_+_23604092 | 0.97 |
ENSDART00000101406
|
rab39bb
|
RAB39B, member RAS oncogene family b |
| chr7_-_25816498 | 0.97 |
ENSDART00000052989
ENSDART00000047951 |
ache
|
acetylcholinesterase |
| chr20_+_48971948 | 0.96 |
ENSDART00000164006
|
nkx2.4b
|
NK2 homeobox 4b |
| chr12_+_4821529 | 0.96 |
|
|
|
| chr24_-_21353742 | 0.95 |
ENSDART00000153695
|
atp8a2
|
ATPase, aminophospholipid transporter, class I, type 8A, member 2 |
| chr9_-_561224 | 0.95 |
|
|
|
| chr1_+_6995893 | 0.94 |
ENSDART00000163488
|
en1b
|
engrailed homeobox 1b |
| chr12_-_552235 | 0.94 |
ENSDART00000168586
ENSDART00000158355 |
bsk146
|
brain specific kinase 146 |
| chr7_-_25426346 | 0.94 |
ENSDART00000082620
|
dysf
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive) |
| chr2_+_7054686 | 0.94 |
ENSDART00000057385
|
nos1apb
|
nitric oxide synthase 1 (neuronal) adaptor protein b |
| chr16_+_5470419 | 0.93 |
|
|
|
| chr10_-_44527708 | 0.93 |
|
|
|
| chr7_-_12715948 | 0.93 |
ENSDART00000173115
|
rplp2l
|
ribosomal protein, large P2, like |
| chr16_+_28449134 | 0.93 |
ENSDART00000059044
|
itga8
|
integrin, alpha 8 |
| chr15_+_16961487 | 0.93 |
ENSDART00000154679
|
ypel2b
|
yippee-like 2b |
| chr12_-_7902815 | 0.92 |
ENSDART00000088100
|
ank3b
|
ankyrin 3b |
| chr10_+_38582701 | 0.92 |
ENSDART00000144329
|
acer3
|
alkaline ceramidase 3 |
| chr6_+_4379989 | 0.92 |
ENSDART00000025031
|
pou4f1
|
POU class 4 homeobox 1 |
| chr17_-_32683561 | 0.90 |
ENSDART00000155002
|
cgref1
|
cell growth regulator with EF-hand domain 1 |
| KN150680v1_+_10854 | 0.90 |
|
|
|
| chr24_+_17112303 | 0.90 |
ENSDART00000131577
|
commd3
|
COMM domain containing 3 |
| chr11_+_6809190 | 0.89 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr23_-_20402258 | 0.89 |
ENSDART00000136204
|
CR749762.2
|
ENSDARG00000093223 |
| chr3_-_56474209 | 0.88 |
ENSDART00000156398
|
si:ch211-189a21.1
|
si:ch211-189a21.1 |
| chr24_+_9605832 | 0.88 |
ENSDART00000131891
|
tmem108
|
transmembrane protein 108 |
| chr1_-_57300743 | 0.88 |
ENSDART00000081122
|
COLGALT1 (1 of many)
|
collagen beta(1-O)galactosyltransferase 1 |
| chr11_+_29913942 | 0.88 |
ENSDART00000157385
|
cdkl5
|
cyclin-dependent kinase-like 5 |
| chr18_+_9228336 | 0.88 |
|
|
|
| chr15_+_3296905 | 0.87 |
ENSDART00000171723
|
foxo1a
|
forkhead box O1 a |
| chr17_-_32683664 | 0.87 |
ENSDART00000109927
|
cgref1
|
cell growth regulator with EF-hand domain 1 |
| chr6_-_39311594 | 0.87 |
|
|
|
| chr11_+_6754335 | 0.87 |
ENSDART00000104292
|
pde4cb
|
phosphodiesterase 4C, cAMP-specific b |
| chr22_+_31866783 | 0.87 |
ENSDART00000159825
|
dock3
|
dedicator of cytokinesis 3 |
| chr20_-_20634087 | 0.87 |
ENSDART00000125039
|
six6b
|
SIX homeobox 6b |
| chr25_-_8475902 | 0.86 |
ENSDART00000176751
|
CU929451.3
|
ENSDARG00000107886 |
| chr9_-_47957181 | 0.86 |
ENSDART00000015159
|
tns1b
|
tensin 1b |
| chr4_-_37424443 | 0.86 |
|
|
|
| chr2_+_35745559 | 0.86 |
ENSDART00000002094
|
ankrd45
|
ankyrin repeat domain 45 |
| chr3_+_17210396 | 0.85 |
ENSDART00000090676
|
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr14_-_16815986 | 0.85 |
ENSDART00000060479
|
smtnl1
|
smoothelin-like 1 |
| chr7_+_36213100 | 0.85 |
ENSDART00000173595
|
chd9
|
chromodomain helicase DNA binding protein 9 |
| chr19_+_156649 | 0.84 |
ENSDART00000167717
|
lrrc16a
|
leucine rich repeat containing 16A |
| chr1_+_2074153 | 0.84 |
ENSDART00000058877
|
rap2ab
|
RAP2A, member of RAS oncogene family b |
| chr17_-_37266525 | 0.84 |
ENSDART00000128715
|
kif3cb
|
kinesin family member 3Cb |
| chr18_-_8697347 | 0.84 |
ENSDART00000093131
|
FRMD4A
|
FERM domain containing 4A |
| chr21_-_38534139 | 0.84 |
ENSDART00000148939
|
pof1b
|
premature ovarian failure, 1B |
| chr21_-_22435721 | 0.84 |
|
|
|
| chr9_-_12063411 | 0.84 |
ENSDART00000038651
|
znf804a
|
zinc finger protein 804A |
| chr24_+_37834148 | 0.83 |
|
|
|
| chr14_-_16762762 | 0.83 |
ENSDART00000106333
|
phox2bb
|
paired-like homeobox 2bb |
| chr6_-_39315608 | 0.83 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr6_+_60152913 | 0.83 |
|
|
|
| chr20_-_35938683 | 0.83 |
ENSDART00000037855
|
tnfrsf21
|
tumor necrosis factor receptor superfamily, member 21 |
| chr19_-_27730419 | 0.82 |
ENSDART00000151047
|
sh3bp5lb
|
SH3-binding domain protein 5-like, b |
| chr17_+_53162459 | 0.82 |
|
|
|
| chr19_-_22904687 | 0.82 |
ENSDART00000141503
|
pleca
|
plectin a |
| chr6_-_43094573 | 0.82 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr9_-_23406315 | 0.82 |
ENSDART00000159256
|
kif5c
|
kinesin family member 5C |
| chr8_-_15267816 | 0.82 |
ENSDART00000137923
|
spata6
|
spermatogenesis associated 6 |
| chr16_+_21098948 | 0.82 |
ENSDART00000006043
|
hoxa11b
|
homeobox A11b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0006581 | response to amphetamine(GO:0001975) acetylcholine catabolic process(GO:0006581) response to amine(GO:0014075) |
| 1.1 | 3.2 | GO:0051101 | regulation of DNA binding(GO:0051101) |
| 0.9 | 2.8 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.8 | 3.1 | GO:0019543 | propionate metabolic process(GO:0019541) propionate catabolic process(GO:0019543) glycerol biosynthetic process from pyruvate(GO:0046327) cellular response to dexamethasone stimulus(GO:0071549) |
| 0.7 | 2.2 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.7 | 3.5 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.7 | 2.1 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.7 | 4.0 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.6 | 2.3 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.5 | 1.5 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.5 | 1.4 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.4 | 1.3 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.4 | 1.2 | GO:0051580 | neurotransmitter uptake(GO:0001504) regulation of neurotransmitter uptake(GO:0051580) neurotransmitter reuptake(GO:0098810) |
| 0.4 | 1.1 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.4 | 2.2 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.4 | 1.8 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.4 | 1.4 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.4 | 1.1 | GO:0010084 | specification of organ axis polarity(GO:0010084) |
| 0.3 | 2.0 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.3 | 1.3 | GO:0000050 | urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.3 | 0.9 | GO:0050765 | monocyte activation involved in immune response(GO:0002280) negative regulation of phagocytosis(GO:0050765) |
| 0.3 | 3.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.3 | 0.9 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.3 | 0.9 | GO:0048659 | smooth muscle cell proliferation(GO:0048659) |
| 0.3 | 0.8 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.2 | 1.0 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.2 | 1.7 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.2 | 1.2 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.2 | 0.7 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) protein K33-linked deubiquitination(GO:1990168) |
| 0.2 | 0.9 | GO:0099558 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.2 | 1.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.2 | 0.6 | GO:0060220 | blood vessel maturation(GO:0001955) retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.2 | 0.8 | GO:0050687 | negative regulation of response to biotic stimulus(GO:0002832) negative regulation of defense response to virus(GO:0050687) |
| 0.2 | 1.0 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 | 1.0 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.2 | 1.3 | GO:0021627 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.2 | 0.6 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.2 | 0.6 | GO:1902893 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.2 | 1.8 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.2 | 1.6 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.2 | 3.0 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.2 | 1.2 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.2 | 0.8 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) axo-dendritic protein transport(GO:0099640) |
| 0.2 | 0.8 | GO:1900028 | negative regulation of Rac protein signal transduction(GO:0035021) wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.2 | 1.0 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.2 | 0.5 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.2 | 1.1 | GO:0090178 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.2 | 0.3 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.1 | 1.2 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 0.6 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 | 0.1 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.1 | 0.5 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 0.8 | GO:1902946 | protein localization to early endosome(GO:1902946) |
| 0.1 | 1.0 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 2.6 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.0 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 3.5 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.1 | 1.3 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 0.4 | GO:0003245 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 0.4 | GO:0051701 | interaction with host(GO:0051701) |
| 0.1 | 8.1 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 0.8 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.3 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 1.1 | GO:0086001 | cardiac muscle cell action potential(GO:0086001) |
| 0.1 | 0.7 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 0.3 | GO:0030431 | sleep(GO:0030431) |
| 0.1 | 2.2 | GO:0070654 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.1 | 0.5 | GO:0006958 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.8 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 0.9 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 0.3 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.1 | 0.6 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 0.3 | GO:0070544 | histone H3-K4 demethylation(GO:0034720) histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 0.4 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 2.3 | GO:0097581 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.1 | 0.8 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 0.5 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 2.1 | GO:0050679 | positive regulation of epithelial cell proliferation(GO:0050679) |
| 0.1 | 0.3 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 0.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.5 | GO:0048901 | anterior lateral line neuromast development(GO:0048901) anterior lateral line neuromast hair cell differentiation(GO:0048903) |
| 0.1 | 0.7 | GO:0042551 | neuron maturation(GO:0042551) |
| 0.1 | 1.0 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.1 | 0.6 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 1.9 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.1 | 0.8 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 1.0 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 1.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.3 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.4 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 2.2 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.1 | 0.5 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.1 | 0.8 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 0.3 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.6 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 0.4 | GO:0060114 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.1 | 0.3 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 0.2 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.1 | 0.7 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.1 | 0.4 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.1 | 0.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.4 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.1 | 0.4 | GO:1901655 | cellular response to ketone(GO:1901655) |
| 0.1 | 1.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 2.1 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 0.2 | GO:0070861 | regulation of COPII vesicle coating(GO:0003400) regulation of protein exit from endoplasmic reticulum(GO:0070861) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.1 | 0.3 | GO:0035909 | aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.1 | 0.3 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 0.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.2 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 1.0 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.0 | 0.6 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.5 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 2.1 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.3 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.2 | GO:0072149 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.0 | 0.5 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.8 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.3 | GO:1902914 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.7 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.6 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.2 | GO:1901021 | positive regulation of calcium ion transmembrane transporter activity(GO:1901021) positive regulation of calcium ion transmembrane transport(GO:1904427) positive regulation of cation channel activity(GO:2001259) |
| 0.0 | 0.6 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.7 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.6 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 1.3 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.2 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 0.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.9 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) estrogen metabolic process(GO:0008210) |
| 0.0 | 0.6 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.1 | GO:2000136 | cell proliferation involved in heart morphogenesis(GO:0061323) regulation of cell proliferation involved in heart morphogenesis(GO:2000136) |
| 0.0 | 1.2 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.4 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 3.3 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.8 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
| 0.0 | 0.1 | GO:0015822 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 | 0.6 | GO:0051181 | cofactor transport(GO:0051181) |
| 0.0 | 1.1 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.4 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 1.2 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.4 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 1.7 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.5 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.9 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.2 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 0.8 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 1.6 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 1.3 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.9 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.2 | GO:0097306 | cellular response to alcohol(GO:0097306) |
| 0.0 | 0.3 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.0 | 2.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 3.1 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.2 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.2 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 1.9 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.2 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.5 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.6 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.4 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.4 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.7 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.3 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:0072401 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.0 | 1.1 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.9 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.7 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.1 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.2 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.8 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.0 | 0.0 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.5 | GO:0044839 | G2/M transition of mitotic cell cycle(GO:0000086) cell cycle G2/M phase transition(GO:0044839) |
| 0.0 | 0.2 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 0.1 | GO:0048009 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 0.1 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.5 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.7 | GO:0051480 | regulation of cytosolic calcium ion concentration(GO:0051480) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 1.0 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.5 | 3.4 | GO:0005833 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 0.3 | 3.5 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.3 | 1.0 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.3 | 0.8 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.2 | 2.7 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.2 | 1.2 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 1.4 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.2 | 1.6 | GO:0098533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.2 | 0.7 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.2 | 0.7 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 2.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.2 | 0.5 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 0.7 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.5 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.6 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.8 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.3 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.1 | 0.6 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 1.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.6 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.4 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 0.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 4.3 | GO:0005604 | basement membrane(GO:0005604) extracellular matrix component(GO:0044420) |
| 0.1 | 0.3 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.1 | 3.0 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 0.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 5.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.9 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 1.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.4 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.2 | GO:0043679 | axon terminus(GO:0043679) neuron projection terminus(GO:0044306) |
| 0.0 | 3.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.8 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.3 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.9 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 10.3 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 1.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 4.0 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.0 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.6 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.7 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 14.3 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.1 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.8 | GO:0030055 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) cell-substrate junction(GO:0030055) |
| 0.0 | 7.0 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 1.1 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.2 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 0.2 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0004104 | acetylcholinesterase activity(GO:0003990) cholinesterase activity(GO:0004104) |
| 1.2 | 3.5 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 1.1 | 3.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.9 | 2.8 | GO:0047150 | betaine-homocysteine S-methyltransferase activity(GO:0047150) |
| 0.8 | 3.1 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.7 | 2.2 | GO:0102345 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.5 | 3.4 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.4 | 1.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.4 | 1.3 | GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.4 | 4.0 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.4 | 1.1 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.3 | 1.0 | GO:0003874 | 6-pyruvoyltetrahydropterin synthase activity(GO:0003874) |
| 0.3 | 1.0 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.3 | 1.0 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.3 | 8.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 0.8 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.2 | 1.4 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.2 | 1.0 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.2 | 0.9 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.2 | 0.7 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) |
| 0.2 | 1.1 | GO:0016801 | hydrolase activity, acting on ether bonds(GO:0016801) |
| 0.2 | 0.7 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.2 | 0.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.2 | 1.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.2 | 3.0 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.2 | 1.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 2.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.2 | 0.8 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.2 | 1.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.4 | GO:0070699 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.1 | 1.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) GABA receptor activity(GO:0016917) |
| 0.1 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.6 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.6 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 1.1 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.3 | GO:0004776 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.1 | 1.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.7 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.1 | 0.3 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.5 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 1.1 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 1.3 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 0.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 1.5 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 1.6 | GO:0015662 | ATPase activity, coupled to transmembrane movement of ions, phosphorylative mechanism(GO:0015662) |
| 0.1 | 1.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.8 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 1.6 | GO:0042562 | hormone binding(GO:0042562) |
| 0.1 | 1.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.8 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 0.7 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 0.4 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.1 | 1.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 4.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.4 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 1.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 1.1 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.4 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 3.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.3 | GO:0019201 | nucleotide kinase activity(GO:0019201) |
| 0.1 | 0.4 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 0.2 | GO:0052725 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) |
| 0.1 | 0.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 0.3 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 0.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.6 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.3 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.3 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 1.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.8 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 2.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.7 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.9 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.8 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.8 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.4 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 1.3 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 1.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 2.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 14.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0031210 | phosphatidylcholine binding(GO:0031210) quaternary ammonium group binding(GO:0050997) |
| 0.0 | 1.8 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.6 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.6 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.3 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 5.2 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.6 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 2.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.5 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.6 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.3 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 4.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 1.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 0.9 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 1.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.7 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 2.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.3 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.8 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.3 | 3.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 4.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 0.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 1.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 2.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 1.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 1.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 1.0 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.2 | 1.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 3.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.0 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 0.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 0.8 | REACTOME ACETYLCHOLINE BINDING AND DOWNSTREAM EVENTS | Genes involved in Acetylcholine Binding And Downstream Events |
| 0.1 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 0.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 2.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 0.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 1.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 5.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 0.4 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 0.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 0.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.8 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.1 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 1.1 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.8 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 1.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |