Project
DANIO-CODE
Navigation
Downloads
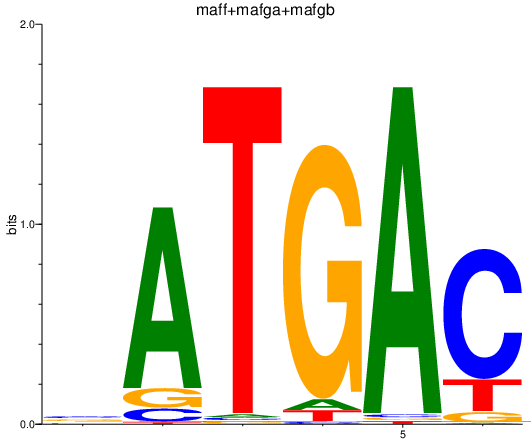
Results for maff+mafga+mafgb
Z-value: 0.73
Transcription factors associated with maff+mafga+mafgb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mafga
|
ENSDARG00000018109 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ga |
|
maff
|
ENSDARG00000028957 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
|
mafgb
|
ENSDARG00000100097 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| maff | dr10_dc_chr12_-_19224698_19224843 | 0.84 | 4.5e-05 | Click! |
| mafgb | dr10_dc_chr11_+_44922098_44922112 | 0.46 | 7.0e-02 | Click! |
Activity profile of maff+mafga+mafgb motif
Sorted Z-values of maff+mafga+mafgb motif
Network of associatons between targets according to the STRING database.
First level regulatory network of maff+mafga+mafgb
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_36487425 | 1.78 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr1_+_25157675 | 1.76 |
ENSDART00000136984
|
fabp2
|
fatty acid binding protein 2, intestinal |
| chr19_-_19888074 | 1.51 |
|
|
|
| chr3_+_41505742 | 1.33 |
ENSDART00000050332
|
gna12a
|
guanine nucleotide binding protein (G protein) alpha 12a |
| chr19_+_17926983 | 1.29 |
ENSDART00000141397
|
ube2e1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr5_+_62611688 | 1.28 |
ENSDART00000177108
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr7_+_26438049 | 1.22 |
ENSDART00000149426
|
cd82a
|
CD82 molecule a |
| chr12_+_6007990 | 1.19 |
ENSDART00000091868
|
g6pca.2
|
glucose-6-phosphatase a, catalytic subunit, tandem duplicate 2 |
| chr11_-_22211478 | 1.18 |
ENSDART00000065996
|
tmem183a
|
transmembrane protein 183A |
| chr17_+_37985095 | 1.15 |
ENSDART00000075941
|
plekhh1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 |
| chr13_-_33574216 | 1.14 |
ENSDART00000065435
|
cst3
|
cystatin C (amyloid angiopathy and cerebral hemorrhage) |
| chr20_+_26639029 | 1.04 |
ENSDART00000045397
|
stx11b.1
|
syntaxin 11b, tandem duplicate 1 |
| chr3_-_26393945 | 1.02 |
ENSDART00000087118
|
xylt1
|
xylosyltransferase I |
| chr5_-_40707316 | 1.02 |
ENSDART00000161932
|
npr3
|
natriuretic peptide receptor 3 |
| chr23_-_28420470 | 0.97 |
ENSDART00000145072
|
neurod4
|
neuronal differentiation 4 |
| chr5_-_24509021 | 0.96 |
ENSDART00000177338
ENSDART00000177557 ENSDART00000177478 |
emid1
|
EMI domain containing 1 |
| chr4_+_3344998 | 0.92 |
ENSDART00000075320
|
nampta
|
nicotinamide phosphoribosyltransferase a |
| chr25_+_222244 | 0.91 |
ENSDART00000155344
|
ENSDARG00000073905
|
ENSDARG00000073905 |
| chr6_+_3549861 | 0.91 |
ENSDART00000170781
|
phospho2
|
phosphatase, orphan 2 |
| chr13_+_24271932 | 0.89 |
ENSDART00000043002
|
rab1ab
|
RAB1A, member RAS oncogene family b |
| chr7_+_40603904 | 0.89 |
ENSDART00000149395
|
shha
|
sonic hedgehog a |
| chr2_-_30071872 | 0.88 |
ENSDART00000056747
|
shhb
|
sonic hedgehog b |
| chr2_-_30675594 | 0.88 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr14_-_46648563 | 0.88 |
|
|
|
| chr12_-_7572970 | 0.87 |
ENSDART00000158095
|
slc16a9b
|
solute carrier family 16, member 9b |
| chr16_+_42925950 | 0.86 |
ENSDART00000159730
ENSDART00000014956 |
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr21_-_41286846 | 0.86 |
ENSDART00000167339
|
msx2b
|
muscle segment homeobox 2b |
| chr3_-_4395852 | 0.86 |
ENSDART00000158197
|
si:dkey-73p2.2
|
si:dkey-73p2.2 |
| chr4_-_13903253 | 0.85 |
ENSDART00000032805
|
gxylt1b
|
glucoside xylosyltransferase 1b |
| chr2_+_32036450 | 0.83 |
ENSDART00000140776
|
CR391940.1
|
ENSDARG00000091946 |
| chr22_+_10172134 | 0.81 |
ENSDART00000132641
|
pdhb
|
pyruvate dehydrogenase (lipoamide) beta |
| chr3_-_25886553 | 0.80 |
ENSDART00000030890
|
hmox1a
|
heme oxygenase 1a |
| chr24_-_986371 | 0.79 |
ENSDART00000168961
ENSDART00000063151 |
napga
|
N-ethylmaleimide-sensitive factor attachment protein, gamma a |
| chr24_-_16884946 | 0.78 |
ENSDART00000171988
|
ptgdsb.2
|
prostaglandin D2 synthase b, tandem duplicate 2 |
| chr10_-_44441481 | 0.77 |
ENSDART00000160231
|
sbno1
|
strawberry notch homolog 1 (Drosophila) |
| chr7_+_56807833 | 0.76 |
ENSDART00000055956
|
enosf1
|
enolase superfamily member 1 |
| chr24_+_9603813 | 0.75 |
ENSDART00000129656
|
tmem108
|
transmembrane protein 108 |
| chr7_-_52848084 | 0.75 |
ENSDART00000172179
ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr2_-_32521879 | 0.75 |
ENSDART00000056639
|
faim2a
|
Fas apoptotic inhibitory molecule 2a |
| chr1_-_50831155 | 0.75 |
ENSDART00000152719
|
spred2a
|
sprouty-related, EVH1 domain containing 2a |
| chr5_-_62816208 | 0.73 |
ENSDART00000097325
|
c5
|
complement component 5 |
| chr22_-_17652938 | 0.73 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr19_-_17926919 | 0.72 |
ENSDART00000167592
ENSDART00000162383 |
nkiras1
|
NFKB inhibitor interacting Ras-like 1 |
| chr10_+_9325719 | 0.72 |
ENSDART00000064968
|
rasgef1bb
|
RasGEF domain family, member 1Bb |
| chr23_+_44817648 | 0.72 |
ENSDART00000143688
|
dlg4b
|
discs, large homolog 4b (Drosophila) |
| chr12_-_1442655 | 0.72 |
|
|
|
| chr25_-_33670713 | 0.71 |
ENSDART00000125036
|
foxb1a
|
forkhead box B1a |
| chr12_-_4767887 | 0.69 |
ENSDART00000167490
|
mapta
|
microtubule-associated protein tau a |
| chr20_-_47123598 | 0.68 |
ENSDART00000100320
|
dnmt3aa
|
DNA (cytosine-5-)-methyltransferase 3 alpha a |
| chr12_-_1442400 | 0.68 |
|
|
|
| chr9_-_21420106 | 0.64 |
ENSDART00000102147
|
popdc2
|
popeye domain containing 2 |
| chr17_+_12544451 | 0.64 |
ENSDART00000064509
ENSDART00000136830 |
stmn4l
|
stathmin-like 4, like |
| chr3_+_33168814 | 0.64 |
ENSDART00000114023
|
hspb9
|
heat shock protein, alpha-crystallin-related, 9 |
| chr18_-_49121584 | 0.63 |
ENSDART00000174157
|
ENSDARG00000099137
|
ENSDARG00000099137 |
| chr1_-_10423289 | 0.62 |
|
|
|
| chr14_+_4489377 | 0.62 |
ENSDART00000041468
|
ap1ar
|
adaptor-related protein complex 1 associated regulatory protein |
| chr17_-_31642673 | 0.62 |
ENSDART00000030448
|
vsx2
|
visual system homeobox 2 |
| chr7_-_64637051 | 0.61 |
ENSDART00000020456
|
mmp15b
|
matrix metallopeptidase 15b |
| chr11_-_22964065 | 0.60 |
|
|
|
| chr12_-_1442470 | 0.60 |
|
|
|
| chr21_-_36338704 | 0.59 |
ENSDART00000157826
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr25_-_13737344 | 0.59 |
|
|
|
| chr7_+_51883765 | 0.59 |
ENSDART00000174389
|
katnb1
|
katanin p80 (WD repeat containing) subunit B 1 |
| chr5_+_53178362 | 0.58 |
ENSDART00000169565
|
sat2b
|
spermidine/spermine N1-acetyltransferase family member 2b |
| chr2_-_30071815 | 0.58 |
ENSDART00000056747
|
shhb
|
sonic hedgehog b |
| chr8_-_21062262 | 0.57 |
|
|
|
| chr11_-_43331509 | 0.56 |
|
|
|
| chr11_-_39864306 | 0.56 |
ENSDART00000165781
|
fam83e
|
family with sequence similarity 83, member E |
| chr6_-_39661423 | 0.55 |
ENSDART00000110380
|
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr24_-_16878970 | 0.54 |
ENSDART00000106058
ENSDART00000106057 |
ptgdsb.2
|
prostaglandin D2 synthase b, tandem duplicate 2 |
| chr4_-_24298444 | 0.54 |
ENSDART00000077926
ENSDART00000128368 |
celf2
|
cugbp, Elav-like family member 2 |
| chr3_+_24145201 | 0.53 |
ENSDART00000169765
|
pdgfbb
|
platelet-derived growth factor beta polypeptide b |
| chr21_+_34895120 | 0.53 |
ENSDART00000157904
ENSDART00000135806 |
si:dkey-71d15.2
|
si:dkey-71d15.2 |
| chr10_+_29966381 | 0.53 |
ENSDART00000141549
|
hspa8
|
heat shock protein 8 |
| chr11_+_10557530 | 0.53 |
ENSDART00000132365
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr20_+_51388660 | 0.53 |
ENSDART00000153452
|
hsp90ab1
|
heat shock protein 90, alpha (cytosolic), class B member 1 |
| chr20_+_54464026 | 0.53 |
ENSDART00000158810
ENSDART00000161631 ENSDART00000172631 ENSDART00000168924 |
fkbp3
|
FK506 binding protein 3 |
| chr19_-_6856033 | 0.52 |
ENSDART00000170952
|
pvrl2l
|
poliovirus receptor-related 2 like |
| chr18_-_1084561 | 0.52 |
ENSDART00000032392
|
dhdhl
|
dihydrodiol dehydrogenase (dimeric), like |
| chr4_-_18466436 | 0.52 |
ENSDART00000150221
|
sco2
|
SCO2 cytochrome c oxidase assembly protein |
| chr2_+_56935721 | 0.51 |
ENSDART00000168497
|
eef2b
|
eukaryotic translation elongation factor 2b |
| chr10_+_29966252 | 0.51 |
ENSDART00000141549
|
hspa8
|
heat shock protein 8 |
| chr19_-_27730419 | 0.51 |
ENSDART00000151047
|
sh3bp5lb
|
SH3-binding domain protein 5-like, b |
| chr1_+_39844690 | 0.51 |
ENSDART00000122059
|
scoca
|
short coiled-coil protein a |
| chr1_-_21987718 | 0.50 |
ENSDART00000128918
|
fgfbp1b
|
fibroblast growth factor binding protein 1b |
| chr1_-_40208469 | 0.50 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr22_+_1404309 | 0.50 |
ENSDART00000161813
|
zgc:101130
|
zgc:101130 |
| chr21_-_26459113 | 0.50 |
ENSDART00000157255
|
cd248b
|
CD248 molecule, endosialin b |
| chr24_+_2990240 | 0.49 |
|
|
|
| chr5_-_62592627 | 0.49 |
ENSDART00000131274
|
tbcelb
|
tubulin folding cofactor E-like b |
| chr14_-_46648119 | 0.49 |
|
|
|
| chr5_-_40451061 | 0.48 |
ENSDART00000083515
|
pdzd2
|
PDZ domain containing 2 |
| chr21_+_5528438 | 0.47 |
ENSDART00000168158
|
shroom3
|
shroom family member 3 |
| chr6_+_34528904 | 0.47 |
ENSDART00000161249
ENSDART00000166715 ENSDART00000161502 |
lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr1_+_35058017 | 0.47 |
ENSDART00000085051
|
hhip
|
hedgehog interacting protein |
| chr17_+_53224434 | 0.47 |
ENSDART00000166517
|
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr8_-_2447758 | 0.46 |
ENSDART00000101125
|
rpl6
|
ribosomal protein L6 |
| chr7_+_65277288 | 0.46 |
|
|
|
| chr10_-_33678206 | 0.45 |
ENSDART00000128049
|
hunk
|
hormonally up-regulated Neu-associated kinase |
| chr4_+_22754618 | 0.45 |
|
|
|
| chr7_+_50491026 | 0.45 |
ENSDART00000165037
|
si:ch73-380l10.2
|
si:ch73-380l10.2 |
| chr25_-_33670651 | 0.45 |
ENSDART00000125036
|
foxb1a
|
forkhead box B1a |
| chr9_-_40212643 | 0.45 |
ENSDART00000166918
|
IKZF2
|
IKAROS family zinc finger 2 |
| chr15_-_2969420 | 0.44 |
|
|
|
| chr18_-_34194840 | 0.44 |
ENSDART00000015079
ENSDART00000133400 |
slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr21_-_43611307 | 0.44 |
ENSDART00000151030
|
si:ch73-362m14.4
|
si:ch73-362m14.4 |
| chr12_-_10262199 | 0.43 |
ENSDART00000126428
|
mpp2b
|
membrane protein, palmitoylated 2b (MAGUK p55 subfamily member 2) |
| KN150266v1_+_74105 | 0.43 |
|
|
|
| chr12_-_28234131 | 0.42 |
ENSDART00000152998
|
ENSDARG00000078481
|
ENSDARG00000078481 |
| chr14_-_46648350 | 0.42 |
|
|
|
| chr25_+_36872560 | 0.42 |
ENSDART00000163178
|
slc10a3
|
solute carrier family 10, member 3 |
| chr5_-_8568715 | 0.42 |
ENSDART00000099891
|
atp5ib
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit Eb |
| chr14_-_46648801 | 0.42 |
|
|
|
| chr18_+_30391910 | 0.41 |
ENSDART00000158871
|
gse1
|
Gse1 coiled-coil protein |
| chr22_+_19605843 | 0.41 |
ENSDART00000124646
|
rgmd
|
RGM domain family, member D |
| chr9_+_19087329 | 0.41 |
ENSDART00000110457
|
crfb1
|
cytokine receptor family member b1 |
| chr14_+_34207218 | 0.41 |
ENSDART00000135608
|
gabrp
|
gamma-aminobutyric acid (GABA) A receptor, pi |
| chr19_+_24340673 | 0.41 |
|
|
|
| chr3_+_33629940 | 0.40 |
ENSDART00000169337
|
ier2a
|
immediate early response 2a |
| chr24_-_25283218 | 0.40 |
ENSDART00000090010
|
phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr4_-_71856428 | 0.40 |
ENSDART00000158977
|
si:ch73-266f23.1
|
si:ch73-266f23.1 |
| chr8_-_47156727 | 0.40 |
|
|
|
| chr23_+_17856104 | 0.40 |
ENSDART00000154427
|
CR381647.1
|
ENSDARG00000097211 |
| chr21_+_45693978 | 0.40 |
ENSDART00000160324
|
CABZ01102047.1
|
ENSDARG00000099648 |
| chr5_+_18826505 | 0.39 |
|
|
|
| chr2_-_36058327 | 0.39 |
ENSDART00000003550
|
nmnat2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr10_+_29965951 | 0.39 |
ENSDART00000116893
|
SNORD14
|
Small nucleolar RNA SNORD14 |
| chr13_+_49527181 | 0.39 |
ENSDART00000176643
|
CABZ01084653.1
|
ENSDARG00000106801 |
| chr3_+_27477793 | 0.38 |
|
|
|
| chr9_-_30073641 | 0.38 |
|
|
|
| chr2_+_37833161 | 0.38 |
ENSDART00000166352
|
sdr39u1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr2_-_14757989 | 0.38 |
ENSDART00000162816
|
BX510915.1
|
ENSDARG00000099630 |
| chr2_-_37914464 | 0.36 |
ENSDART00000129852
|
hbl1
|
hexose-binding lectin 1 |
| chr3_-_32471212 | 0.36 |
ENSDART00000150997
|
si:ch73-248e21.7
|
si:ch73-248e21.7 |
| chr3_-_54884533 | 0.36 |
ENSDART00000115324
|
hbae1.3
|
hemoglobin, alpha embryonic 1.3 |
| chr25_-_14573221 | 0.36 |
|
|
|
| chr23_+_29431173 | 0.36 |
ENSDART00000027255
|
tardbpl
|
TAR DNA binding protein, like |
| chr8_+_27788830 | 0.36 |
ENSDART00000078509
|
capza1b
|
capping protein (actin filament) muscle Z-line, alpha 1b |
| chr20_+_52740555 | 0.36 |
ENSDART00000110777
|
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr14_+_33073643 | 0.35 |
ENSDART00000052780
|
ndufa1
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1 |
| chr23_-_27418761 | 0.35 |
ENSDART00000022042
|
scn8aa
|
sodium channel, voltage gated, type VIII, alpha subunit a |
| chr17_-_5703094 | 0.35 |
ENSDART00000058894
|
si:ch73-340m8.2
|
si:ch73-340m8.2 |
| chr14_-_25688444 | 0.35 |
ENSDART00000172909
|
atox1
|
antioxidant 1 copper chaperone |
| chr19_+_17832515 | 0.35 |
ENSDART00000149045
|
klf2b
|
Kruppel-like factor 2b |
| KN150708v1_+_30684 | 0.35 |
|
|
|
| chr23_-_36198253 | 0.34 |
|
|
|
| chr22_+_11490729 | 0.34 |
ENSDART00000063147
|
sgsh
|
N-sulfoglucosamine sulfohydrolase (sulfamidase) |
| chr6_+_49998103 | 0.34 |
|
|
|
| chr3_+_31802145 | 0.33 |
ENSDART00000139644
|
lin7b
|
lin-7 homolog B (C. elegans) |
| chr1_+_25717717 | 0.33 |
ENSDART00000112263
|
arhgef38
|
Rho guanine nucleotide exchange factor (GEF) 38 |
| chr19_-_33341287 | 0.33 |
ENSDART00000050750
|
rrm2b
|
ribonucleotide reductase M2 b |
| chr7_+_20377678 | 0.33 |
ENSDART00000173710
|
si:dkey-19b23.15
|
si:dkey-19b23.15 |
| chr21_+_15613081 | 0.33 |
ENSDART00000065772
|
ddt
|
D-dopachrome tautomerase |
| chr10_+_29966533 | 0.33 |
ENSDART00000141549
|
hspa8
|
heat shock protein 8 |
| chr4_+_12617165 | 0.33 |
ENSDART00000003583
|
lmo3
|
LIM domain only 3 |
| chr16_+_24046789 | 0.32 |
|
|
|
| chr5_+_296856 | 0.32 |
|
|
|
| chr25_+_446920 | 0.32 |
ENSDART00000104717
|
rsl24d1
|
ribosomal L24 domain containing 1 |
| chr2_-_30071993 | 0.32 |
ENSDART00000056747
|
shhb
|
sonic hedgehog b |
| chr21_-_9863071 | 0.32 |
ENSDART00000170710
|
arhgap24
|
Rho GTPase activating protein 24 |
| chr20_+_54463948 | 0.32 |
ENSDART00000158810
ENSDART00000161631 ENSDART00000172631 ENSDART00000168924 |
fkbp3
|
FK506 binding protein 3 |
| chr23_-_4979524 | 0.31 |
ENSDART00000060714
|
atp6ap1a
|
ATPase, H+ transporting, lysosomal accessory protein 1a |
| chr19_+_26065401 | 0.31 |
ENSDART00000146947
|
tac1
|
tachykinin 1 |
| chr5_-_24509180 | 0.31 |
ENSDART00000177338
ENSDART00000177557 ENSDART00000177478 |
emid1
|
EMI domain containing 1 |
| chr14_+_51724430 | 0.31 |
ENSDART00000168437
|
b4galt1l
|
DP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1, like |
| KN149755v1_+_2933 | 0.31 |
|
|
|
| chr8_-_31065717 | 0.31 |
ENSDART00000163407
|
slc20a1a
|
solute carrier family 20, member 1a |
| chr25_+_34629404 | 0.30 |
ENSDART00000156376
|
CR762436.2
|
Histone H3.2 |
| chr15_+_2894254 | 0.30 |
ENSDART00000163434
|
mre11a
|
MRE11 homolog A, double strand break repair nuclease |
| chr5_+_37184963 | 0.30 |
ENSDART00000053511
|
myo1ca
|
myosin Ic, paralog a |
| chr14_+_20614087 | 0.30 |
ENSDART00000160318
|
lygl2
|
lysozyme g-like 2 |
| chr19_-_48144752 | 0.30 |
ENSDART00000158979
|
c19h1orf109
|
c19h1orf109 homolog (H. sapiens) |
| chr2_-_27920046 | 0.29 |
ENSDART00000014568
|
urod
|
uroporphyrinogen decarboxylase |
| chr11_+_30482530 | 0.29 |
ENSDART00000103270
|
slc22a7a
|
solute carrier family 22 (organic anion transporter), member 7a |
| chr14_-_952510 | 0.29 |
ENSDART00000130801
|
acsl1b
|
acyl-CoA synthetase long-chain family member 1b |
| chr17_-_37247505 | 0.29 |
ENSDART00000108514
|
asxl2
|
additional sex combs like transcriptional regulator 2 |
| chr7_+_36267647 | 0.29 |
ENSDART00000173653
|
chd9
|
chromodomain helicase DNA binding protein 9 |
| chr12_-_1442212 | 0.28 |
|
|
|
| chr22_-_11106940 | 0.28 |
ENSDART00000016873
|
atp6ap2
|
ATPase, H+ transporting, lysosomal accessory protein 2 |
| chr3_+_24564249 | 0.28 |
ENSDART00000111769
|
mkl1a
|
megakaryoblastic leukemia (translocation) 1a |
| chr8_-_46903869 | 0.27 |
ENSDART00000161462
|
acot7
|
acyl-CoA thioesterase 7 |
| chr15_-_5575675 | 0.27 |
ENSDART00000099520
|
atg16l2
|
ATG16 autophagy related 16-like 2 (S. cerevisiae) |
| chr25_+_34546629 | 0.27 |
ENSDART00000133379
|
hist2h3c
|
histone cluster 2, H3c |
| chr19_-_2011177 | 0.27 |
ENSDART00000108784
|
mturn
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr5_+_28170113 | 0.27 |
ENSDART00000114473
|
trafd1
|
TRAF-type zinc finger domain containing 1 |
| chr10_-_17264097 | 0.27 |
ENSDART00000134059
|
depdc5
|
DEP domain containing 5 |
| chr5_-_60747054 | 0.26 |
ENSDART00000082952
|
rcc1l
|
RCC1 like |
| chr10_+_37324880 | 0.26 |
ENSDART00000169027
|
nf1b
|
neurofibromin 1b |
| chr4_+_59014576 | 0.26 |
|
|
|
| chr15_-_37832225 | 0.26 |
ENSDART00000059568
|
si:ch211-137j23.6
|
si:ch211-137j23.6 |
| chr20_+_27494571 | 0.26 |
ENSDART00000005473
|
tmem179
|
transmembrane protein 179 |
| chr4_+_33077499 | 0.26 |
|
|
|
| chr7_-_15004167 | 0.26 |
ENSDART00000030009
|
ENSDARG00000023868
|
ENSDARG00000023868 |
| chr8_+_28433869 | 0.24 |
ENSDART00000062716
|
slc52a3
|
solute carrier family 52 (riboflavin transporter), member 3 |
| chr16_-_7876036 | 0.24 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr17_-_5453357 | 0.24 |
ENSDART00000004043
|
enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr10_+_45213266 | 0.24 |
ENSDART00000166945
|
nudcd3
|
NudC domain containing 3 |
| chr21_+_27379622 | 0.24 |
ENSDART00000108763
|
cfb
|
complement factor B |
| chr8_+_45353831 | 0.24 |
ENSDART00000149418
|
chmp4bb
|
charged multivesicular body protein 4Bb |
| chr20_+_51388752 | 0.24 |
ENSDART00000153452
|
hsp90ab1
|
heat shock protein 90, alpha (cytosolic), class B member 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0061373 | associative learning(GO:0008306) visual learning(GO:0008542) mammillary axonal complex development(GO:0061373) mammillothalamic axonal tract development(GO:0061374) corpora quadrigemina development(GO:0061378) inferior colliculus development(GO:0061379) cell migration in diencephalon(GO:0061381) |
| 0.4 | 0.8 | GO:0003379 | establishment of cell polarity involved in gastrulation cell migration(GO:0003379) |
| 0.3 | 0.9 | GO:1905178 | regulation of cardiac muscle tissue regeneration(GO:1905178) |
| 0.3 | 0.8 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.3 | 0.8 | GO:0007585 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.3 | 0.8 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.2 | 0.7 | GO:0072578 | neurotransmitter-gated ion channel clustering(GO:0072578) |
| 0.2 | 1.4 | GO:1902946 | protein localization to early endosome(GO:1902946) |
| 0.2 | 0.9 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.2 | 1.8 | GO:0060956 | cardiac endothelial cell differentiation(GO:0003348) endocardial cell differentiation(GO:0060956) |
| 0.2 | 0.8 | GO:0036363 | transforming growth factor beta activation(GO:0036363) transforming growth factor beta production(GO:0071604) regulation of transforming growth factor beta production(GO:0071634) negative regulation of transforming growth factor beta production(GO:0071635) regulation of transforming growth factor beta activation(GO:1901388) negative regulation of transforming growth factor beta activation(GO:1901389) |
| 0.2 | 0.7 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.2 | 1.3 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.1 | 0.8 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.5 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 0.1 | 0.9 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.1 | 0.6 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.1 | 1.0 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.1 | 0.3 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.1 | 0.4 | GO:0030194 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.1 | 0.7 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.1 | 0.6 | GO:1900024 | regulation of substrate adhesion-dependent cell spreading(GO:1900024) |
| 0.1 | 0.3 | GO:0044036 | cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.1 | 0.5 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.1 | 0.5 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.2 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.1 | 0.5 | GO:0044034 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.6 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.9 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.1 | 0.5 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.3 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.2 | GO:0009215 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) purine deoxyribonucleoside triphosphate metabolic process(GO:0009215) |
| 0.1 | 1.2 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 0.5 | GO:0060251 | regulation of glial cell proliferation(GO:0060251) |
| 0.1 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 1.0 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 | 0.2 | GO:0009838 | abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) |
| 0.1 | 0.2 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 0.3 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 | 1.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.3 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.1 | 0.6 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.1 | 0.7 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.1 | 1.0 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.1 | 0.3 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.1 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.3 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.6 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.6 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.0 | 1.1 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.0 | 0.4 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.4 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.5 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.1 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.0 | 0.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0086001 | cardiac muscle cell action potential(GO:0086001) |
| 0.0 | 0.9 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.1 | GO:0060911 | cardiac cell fate commitment(GO:0060911) |
| 0.0 | 0.3 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.2 | GO:0006956 | complement activation(GO:0006956) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.1 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.6 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.3 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.5 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.7 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.5 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.3 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.1 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 | 0.4 | GO:0021955 | central nervous system neuron axonogenesis(GO:0021955) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.0 | 1.8 | GO:0033993 | response to lipid(GO:0033993) |
| 0.0 | 0.3 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.6 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.6 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.4 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.4 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 0.2 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 0.3 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.1 | GO:0051965 | regulation of synapse assembly(GO:0051963) positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.8 | GO:0009063 | cellular amino acid catabolic process(GO:0009063) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.3 | 1.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 0.6 | GO:0008352 | katanin complex(GO:0008352) |
| 0.1 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 0.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 1.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.4 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 0.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 0.7 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 0.4 | GO:0005833 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.3 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 0.8 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.1 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.8 | GO:0070161 | adherens junction(GO:0005912) anchoring junction(GO:0070161) |
| 0.0 | 0.1 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.0 | 0.6 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.8 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.3 | 2.7 | GO:0005113 | patched binding(GO:0005113) |
| 0.2 | 0.9 | GO:0019808 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.2 | 1.3 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.2 | 0.8 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) |
| 0.2 | 0.8 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.2 | 0.5 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 1.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.4 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.1 | 0.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.8 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 1.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.3 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.3 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.1 | 0.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.4 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 0.3 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 0.4 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.4 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.4 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.3 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.7 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 1.0 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.3 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.3 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 1.1 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.0 | 0.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.0 | 0.1 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.0 | 0.1 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 0.5 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.3 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.4 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.5 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.8 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.0 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.8 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 1.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.7 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 1.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.8 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 0.8 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 1.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 0.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 0.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.5 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 1.3 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 1.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |