Project
DANIO-CODE
Navigation
Downloads
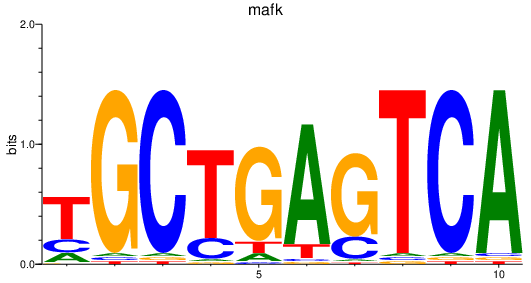
Results for mafk
Z-value: 1.82
Transcription factors associated with mafk
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mafk
|
ENSDARG00000100947 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mafk | dr10_dc_chr3_-_42275696_42275807 | 0.85 | 2.9e-05 | Click! |
Activity profile of mafk motif
Sorted Z-values of mafk motif
Network of associatons between targets according to the STRING database.
First level regulatory network of mafk
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_25886553 | 7.83 |
ENSDART00000030890
|
hmox1a
|
heme oxygenase 1a |
| chr3_-_32686790 | 7.06 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr16_-_9978112 | 5.72 |
ENSDART00000149312
|
ncalda
|
neurocalcin delta a |
| chr5_-_66792947 | 4.90 |
ENSDART00000147009
|
si:dkey-251i10.2
|
si:dkey-251i10.2 |
| chr11_+_26371444 | 4.78 |
ENSDART00000042322
|
map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr19_-_7501777 | 4.75 |
ENSDART00000081741
|
rab25a
|
RAB25, member RAS oncogene family a |
| chr6_-_39661423 | 4.72 |
ENSDART00000110380
|
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr8_-_17480730 | 4.53 |
ENSDART00000100667
|
skia
|
v-ski avian sarcoma viral oncogene homolog a |
| chr20_+_34423970 | 4.16 |
ENSDART00000061659
|
ivns1abpa
|
influenza virus NS1A binding protein a |
| chr14_+_11151485 | 4.04 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr5_+_42312784 | 4.00 |
ENSDART00000039973
|
rufy3
|
RUN and FYVE domain containing 3 |
| chr9_+_21581061 | 3.90 |
|
|
|
| chr4_-_2615160 | 3.52 |
ENSDART00000140760
|
e2f7
|
E2F transcription factor 7 |
| chr5_-_30015572 | 3.32 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr15_-_24987099 | 3.31 |
|
|
|
| chr16_-_16274298 | 3.30 |
ENSDART00000103815
|
stmn2a
|
stathmin 2a |
| chr5_+_36487425 | 3.30 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr9_-_24431684 | 3.23 |
ENSDART00000039399
|
cavin2a
|
caveolae associated protein 2a |
| chr13_+_2774422 | 3.22 |
ENSDART00000162362
|
wu:fj16a03
|
wu:fj16a03 |
| chr23_+_19887319 | 3.20 |
ENSDART00000139192
|
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr10_+_38831967 | 3.18 |
ENSDART00000125045
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr2_-_1677190 | 3.17 |
ENSDART00000024135
|
tubb2
|
tubulin, beta 2A class IIa |
| chr11_-_2107054 | 3.10 |
ENSDART00000173031
|
hoxc6b
|
homeobox C6b |
| chr19_+_32183937 | 3.09 |
ENSDART00000147474
|
stmn2b
|
stathmin 2b |
| chr22_+_21997000 | 3.04 |
ENSDART00000046174
|
gna15.1
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 1 |
| chr5_+_65267043 | 3.00 |
ENSDART00000161578
|
mymk
|
myomaker, myoblast fusion factor |
| chr20_-_39200252 | 2.96 |
ENSDART00000037318
ENSDART00000143379 |
rcan2
|
regulator of calcineurin 2 |
| chr7_+_23843682 | 2.90 |
ENSDART00000173899
|
mrpl52
|
mitochondrial ribosomal protein L52 |
| chr23_+_26215871 | 2.86 |
ENSDART00000158878
|
ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 |
| chr23_+_10412852 | 2.86 |
ENSDART00000142595
|
krt18
|
keratin 18 |
| chr14_+_34440699 | 2.81 |
ENSDART00000130469
|
sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr15_+_29343644 | 2.79 |
ENSDART00000170537
ENSDART00000128973 |
rap1gap2a
|
RAP1 GTPase activating protein 2a |
| chr12_-_1931281 | 2.75 |
ENSDART00000005676
ENSDART00000127937 |
sox9a
|
SRY (sex determining region Y)-box 9a |
| chr5_+_64225977 | 2.72 |
ENSDART00000122863
|
ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr1_-_51862897 | 2.64 |
ENSDART00000136469
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr21_-_41001910 | 2.60 |
ENSDART00000142277
|
plac8l1
|
PLAC8-like 1 |
| chr9_+_41301317 | 2.59 |
ENSDART00000000250
|
slc40a1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr21_+_30757831 | 2.55 |
ENSDART00000139486
|
ENSDARG00000030006
|
ENSDARG00000030006 |
| chr24_+_30343717 | 2.52 |
ENSDART00000162377
|
FP085414.1
|
ENSDARG00000100270 |
| chr3_+_25933164 | 2.51 |
ENSDART00000143697
|
si:dkeyp-69e1.8
|
si:dkeyp-69e1.8 |
| chr12_+_38382027 | 2.50 |
ENSDART00000021069
|
rpl38
|
ribosomal protein L38 |
| chr11_-_32461160 | 2.48 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr9_-_3700395 | 2.44 |
ENSDART00000102900
|
sp5a
|
Sp5 transcription factor a |
| chr23_-_27645138 | 2.43 |
ENSDART00000008174
|
pfkma
|
phosphofructokinase, muscle a |
| chr7_+_56807833 | 2.43 |
ENSDART00000055956
|
enosf1
|
enolase superfamily member 1 |
| chr1_-_7456961 | 2.42 |
ENSDART00000152295
|
FAM83G
|
family with sequence similarity 83 member G |
| chr5_+_64226125 | 2.39 |
ENSDART00000122863
|
ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr19_-_23037220 | 2.38 |
ENSDART00000090669
|
pleca
|
plectin a |
| chr16_+_21109486 | 2.37 |
ENSDART00000079383
|
hoxa9b
|
homeobox A9b |
| chr18_-_17031173 | 2.33 |
ENSDART00000129146
|
tbc1d15
|
TBC1 domain family, member 15 |
| chr14_+_34440799 | 2.33 |
ENSDART00000173371
|
sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr10_+_38831925 | 2.30 |
ENSDART00000125045
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr14_+_26137608 | 2.28 |
ENSDART00000175459
|
gpr137
|
G protein-coupled receptor 137 |
| chr21_-_23074223 | 2.27 |
ENSDART00000147896
|
usp28
|
ubiquitin specific peptidase 28 |
| chr19_+_31817286 | 2.25 |
ENSDART00000078459
|
tmem55a
|
transmembrane protein 55A |
| chr16_-_17254837 | 2.24 |
ENSDART00000089386
|
iffo1b
|
intermediate filament family orphan 1b |
| chr25_-_22089794 | 2.24 |
ENSDART00000139110
|
pkp3a
|
plakophilin 3a |
| chr14_+_24543732 | 2.22 |
ENSDART00000106039
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr13_-_22877318 | 2.21 |
ENSDART00000057638
ENSDART00000171778 |
hk1
|
hexokinase 1 |
| chr1_-_25272318 | 2.20 |
ENSDART00000134192
|
synpo2b
|
synaptopodin 2b |
| chr11_-_39864306 | 2.19 |
ENSDART00000165781
|
fam83e
|
family with sequence similarity 83, member E |
| chr15_-_40391882 | 2.16 |
|
|
|
| chr9_-_23307419 | 2.16 |
ENSDART00000020884
|
lypd6
|
LY6/PLAUR domain containing 6 |
| chr7_+_20046425 | 2.08 |
ENSDART00000131019
|
acadvl
|
acyl-CoA dehydrogenase, very long chain |
| chr12_-_4648262 | 2.07 |
ENSDART00000152771
|
si:ch211-255p10.3
|
si:ch211-255p10.3 |
| chr16_+_11138924 | 2.06 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr19_-_41819752 | 2.05 |
ENSDART00000167772
|
shfm1
|
split hand/foot malformation (ectrodactyly) type 1 |
| chr3_-_4395852 | 2.02 |
ENSDART00000158197
|
si:dkey-73p2.2
|
si:dkey-73p2.2 |
| chr13_-_35908743 | 2.02 |
ENSDART00000133565
|
si:dkey-157l19.2
|
si:dkey-157l19.2 |
| chr3_+_25992836 | 1.97 |
ENSDART00000010477
|
hsp70.3
|
heat shock cognate 70-kd protein, tandem duplicate 3 |
| chr20_+_51385187 | 1.97 |
|
|
|
| chr16_+_11138879 | 1.95 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr5_-_37335445 | 1.94 |
ENSDART00000141233
|
scn4bb
|
sodium channel, voltage-gated, type IV, beta b |
| chr6_-_34951506 | 1.93 |
|
|
|
| chr23_+_26995808 | 1.93 |
|
|
|
| chr3_-_21149752 | 1.92 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr10_+_31336284 | 1.86 |
ENSDART00000140988
|
tmem218
|
transmembrane protein 218 |
| chr14_-_31522323 | 1.84 |
ENSDART00000172870
|
rbmx
|
RNA binding motif protein, X-linked |
| chr13_+_28689749 | 1.84 |
ENSDART00000101653
|
CU639469.1
|
ENSDARG00000062790 |
| chr8_-_15071283 | 1.83 |
|
|
|
| chr20_-_25727145 | 1.83 |
ENSDART00000126716
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr23_+_14058972 | 1.83 |
ENSDART00000090864
|
lmod3
|
leiomodin 3 (fetal) |
| chr2_-_25325686 | 1.82 |
ENSDART00000111212
|
nck1a
|
NCK adaptor protein 1a |
| chr10_+_26786051 | 1.81 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr24_+_9073790 | 1.81 |
|
|
|
| chr22_+_520496 | 1.79 |
ENSDART00000067638
|
med20
|
mediator complex subunit 20 |
| chr2_+_10023967 | 1.76 |
ENSDART00000148227
|
anxa13l
|
annexin A13, like |
| chr14_-_49600662 | 1.76 |
ENSDART00000161147
|
si:ch211-154c21.1
|
si:ch211-154c21.1 |
| chr6_-_34951317 | 1.76 |
|
|
|
| chr12_-_28248133 | 1.74 |
ENSDART00000016283
|
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr6_+_12618821 | 1.72 |
ENSDART00000156341
|
fam117ba
|
family with sequence similarity 117, member Ba |
| chr12_+_2412047 | 1.71 |
ENSDART00000112032
|
ARHGAP22 (1 of many)
|
Rho GTPase activating protein 22 |
| chr8_-_50270783 | 1.71 |
ENSDART00000146056
|
nkx3-1
|
NK3 homeobox 1 |
| chr14_-_17282615 | 1.70 |
ENSDART00000006716
ENSDART00000136242 |
selt2
|
selenoprotein T, 2 |
| chr14_+_47229400 | 1.70 |
ENSDART00000136045
|
mgst2
|
microsomal glutathione S-transferase 2 |
| chr21_+_5887486 | 1.70 |
ENSDART00000048399
|
slc4a4b
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4b |
| chr24_-_37792982 | 1.69 |
ENSDART00000078828
ENSDART00000131342 |
anks3
|
ankyrin repeat and sterile alpha motif domain containing 3 |
| chr1_+_50395721 | 1.68 |
ENSDART00000134065
|
dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr10_+_9764591 | 1.67 |
ENSDART00000091780
|
rc3h2
|
ring finger and CCCH-type domains 2 |
| chr22_-_26728956 | 1.66 |
ENSDART00000169887
|
wu:fu71h07
|
wu:fu71h07 |
| chr3_-_17881044 | 1.66 |
ENSDART00000013540
|
si:ch73-141c7.1
|
si:ch73-141c7.1 |
| chr7_+_10346939 | 1.66 |
|
|
|
| chr17_+_45430353 | 1.64 |
ENSDART00000162937
|
ezra
|
ezrin a |
| chr22_+_37696217 | 1.64 |
ENSDART00000007346
|
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr17_+_27383737 | 1.64 |
ENSDART00000156756
|
FP017274.1
|
ENSDARG00000097369 |
| chr7_+_39116005 | 1.62 |
ENSDART00000146702
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr22_-_13325475 | 1.62 |
ENSDART00000154095
|
si:ch211-227m13.1
|
si:ch211-227m13.1 |
| chr22_+_28496424 | 1.60 |
ENSDART00000089546
|
abi3bpb
|
ABI family, member 3 (NESH) binding protein b |
| chr3_+_21059221 | 1.60 |
ENSDART00000078807
|
zgc:123295
|
zgc:123295 |
| chr6_-_39656043 | 1.58 |
ENSDART00000155859
|
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr11_-_39864543 | 1.58 |
ENSDART00000165781
|
fam83e
|
family with sequence similarity 83, member E |
| chr4_-_376986 | 1.58 |
ENSDART00000067482
ENSDART00000160718 |
dynlt1
|
dynein, light chain, Tctex-type 1 |
| chr21_-_35646193 | 1.57 |
|
|
|
| chr4_+_13569588 | 1.56 |
ENSDART00000136152
|
calua
|
calumenin a |
| chr24_+_18154733 | 1.54 |
ENSDART00000140994
|
tpk1
|
thiamin pyrophosphokinase 1 |
| chr3_+_49166063 | 1.53 |
ENSDART00000156347
|
epn3a
|
epsin 3a |
| chr9_+_36316158 | 1.52 |
ENSDART00000176763
|
lrp1bb
|
low density lipoprotein receptor-related protein 1Bb |
| chr7_+_16615075 | 1.51 |
|
|
|
| chr2_+_19587617 | 1.50 |
ENSDART00000166292
|
cc2d1b
|
coiled-coil and C2 domain containing 1B |
| chr21_+_9483423 | 1.49 |
ENSDART00000162834
|
mapk10
|
mitogen-activated protein kinase 10 |
| chr1_+_13386937 | 1.49 |
ENSDART00000021693
|
ank2a
|
ankyrin 2a, neuronal |
| chr24_+_14093524 | 1.49 |
|
|
|
| chr20_+_29307142 | 1.49 |
ENSDART00000153016
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr11_-_25181234 | 1.49 |
ENSDART00000013714
|
gata1a
|
GATA binding protein 1a |
| chr5_+_26925392 | 1.48 |
ENSDART00000051491
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr2_+_52473102 | 1.47 |
ENSDART00000146418
|
shda
|
Src homology 2 domain containing transforming protein D, a |
| chr24_+_26922969 | 1.46 |
|
|
|
| chr8_-_50299273 | 1.41 |
ENSDART00000023639
|
nkx2.7
|
NK2 transcription factor related 7 |
| chr5_-_18548689 | 1.41 |
ENSDART00000137802
|
fam214b
|
family with sequence similarity 214, member B |
| chr13_-_11553609 | 1.40 |
|
|
|
| chr7_+_72128846 | 1.40 |
ENSDART00000123887
|
MAPK8IP1 (1 of many)
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr2_+_25622497 | 1.39 |
ENSDART00000131977
|
ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr21_+_45780315 | 1.38 |
|
|
|
| chr18_+_29178482 | 1.38 |
ENSDART00000137587
ENSDART00000144423 |
ppfibp2a
|
PTPRF interacting protein, binding protein 2a (liprin beta 2) |
| chr20_+_29307039 | 1.37 |
ENSDART00000152949
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr24_+_30343687 | 1.36 |
ENSDART00000162377
|
FP085414.1
|
ENSDARG00000100270 |
| chr3_-_28120668 | 1.34 |
|
|
|
| chr16_-_21981065 | 1.33 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr2_-_26987186 | 1.33 |
ENSDART00000132854
|
u2surp
|
U2 snRNP-associated SURP domain containing |
| chr20_-_1293519 | 1.32 |
ENSDART00000152436
|
pcmt
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr1_+_18610282 | 1.31 |
ENSDART00000129970
|
si:dkeyp-118a3.2
|
si:dkeyp-118a3.2 |
| chr11_+_18020191 | 1.29 |
ENSDART00000125984
|
mustn1b
|
musculoskeletal, embryonic nuclear protein 1b |
| chr19_-_7235125 | 1.28 |
|
|
|
| chr18_+_14369543 | 1.28 |
|
|
|
| chr6_-_120376 | 1.26 |
ENSDART00000148974
|
keap1b
|
kelch-like ECH-associated protein 1b |
| chr16_-_19154920 | 1.26 |
ENSDART00000088818
|
fhod3b
|
formin homology 2 domain containing 3b |
| chr22_-_4622401 | 1.25 |
ENSDART00000106166
|
rx1
|
retinal homeobox gene 1 |
| chr15_-_19186046 | 1.25 |
ENSDART00000108818
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr10_-_28474085 | 1.24 |
|
|
|
| chr24_-_1308951 | 1.24 |
|
|
|
| chr23_+_14461509 | 1.24 |
ENSDART00000143618
|
birc7
|
baculoviral IAP repeat containing 7 |
| chr1_+_15827083 | 1.22 |
|
|
|
| chr18_+_45564871 | 1.21 |
|
|
|
| chr6_-_39655998 | 1.19 |
ENSDART00000155859
|
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr3_-_30027552 | 1.19 |
ENSDART00000103502
|
si:ch211-152f23.5
|
si:ch211-152f23.5 |
| chr24_+_16760759 | 1.18 |
ENSDART00000066760
|
cct5
|
chaperonin containing TCP1, subunit 5 (epsilon) |
| chr6_+_12619062 | 1.17 |
ENSDART00000156341
|
fam117ba
|
family with sequence similarity 117, member Ba |
| chr16_+_13964949 | 1.17 |
ENSDART00000143983
|
zgc:174888
|
zgc:174888 |
| chr3_-_12818954 | 1.17 |
ENSDART00000158747
ENSDART00000158815 |
pdgfab
|
platelet-derived growth factor alpha polypeptide b |
| chr23_+_5632124 | 1.17 |
ENSDART00000059307
|
smpd2a
|
sphingomyelin phosphodiesterase 2a, neutral membrane (neutral sphingomyelinase) |
| chr1_-_39509011 | 1.14 |
ENSDART00000146680
|
ENSDARG00000078251
|
ENSDARG00000078251 |
| chr25_-_15117843 | 1.14 |
ENSDART00000031499
|
wt1a
|
wilms tumor 1a |
| chr24_-_37793048 | 1.14 |
ENSDART00000078828
ENSDART00000131342 |
anks3
|
ankyrin repeat and sterile alpha motif domain containing 3 |
| chr12_+_35912088 | 1.12 |
ENSDART00000167873
|
baiap2b
|
BAI1-associated protein 2b |
| chr18_+_22228682 | 1.12 |
ENSDART00000165464
|
fam65a
|
family with sequence similarity 65, member A |
| chr24_+_27469268 | 1.11 |
ENSDART00000105774
|
ek1
|
eph-like kinase 1 |
| chr19_-_10962536 | 1.10 |
ENSDART00000160438
|
psmd4a
|
proteasome 26S subunit, non-ATPase 4a |
| chr5_-_44243476 | 1.10 |
ENSDART00000161408
|
fbp1a
|
fructose-1,6-bisphosphatase 1a |
| chr22_-_16971035 | 1.10 |
ENSDART00000090237
|
nfia
|
nuclear factor I/A |
| chr5_-_20527105 | 1.09 |
ENSDART00000133461
|
camk2b1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 1 |
| chr12_-_31342432 | 1.09 |
ENSDART00000148603
|
acsl5
|
acyl-CoA synthetase long-chain family member 5 |
| chr25_-_36886647 | 1.09 |
ENSDART00000160688
|
psma1
|
proteasome subunit alpha 1 |
| chr7_+_620117 | 1.08 |
ENSDART00000152224
|
zgc:63470
|
zgc:63470 |
| chr17_-_14956738 | 1.08 |
|
|
|
| chr2_-_25306278 | 1.08 |
ENSDART00000132050
|
hltf
|
helicase-like transcription factor |
| chr24_+_19842162 | 1.07 |
ENSDART00000123031
|
CR381686.3
|
ENSDARG00000094073 |
| chr5_-_18548376 | 1.05 |
ENSDART00000133330
|
fam214b
|
family with sequence similarity 214, member B |
| chr12_-_28671833 | 1.05 |
|
|
|
| chr19_-_10324632 | 1.05 |
ENSDART00000148073
|
shisa7b
|
shisa family member 7 |
| chr4_-_72146177 | 1.04 |
ENSDART00000150546
|
si:dkey-262g12.3
|
si:dkey-262g12.3 |
| chr20_+_18762031 | 1.04 |
ENSDART00000152136
ENSDART00000126959 |
tnfaip2a
|
tumor necrosis factor, alpha-induced protein 2a |
| chr3_-_28120255 | 1.03 |
|
|
|
| chr4_-_885413 | 1.03 |
ENSDART00000022668
|
aim1b
|
crystallin beta-gamma domain containing 1b |
| chr17_-_35011682 | 1.02 |
|
|
|
| chr2_+_50892294 | 1.02 |
ENSDART00000018150
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr9_-_12063411 | 1.02 |
ENSDART00000038651
|
znf804a
|
zinc finger protein 804A |
| chr17_+_48041768 | 1.00 |
ENSDART00000160219
|
slc39a9
|
solute carrier family 39, member 9 |
| chr3_+_45345181 | 0.98 |
|
|
|
| chr22_-_31110564 | 0.97 |
ENSDART00000022445
|
cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr17_-_12812747 | 0.96 |
ENSDART00000022874
ENSDART00000046025 |
psma6a
|
proteasome subunit alpha 6a |
| chr6_+_31377287 | 0.96 |
|
|
|
| chr23_+_24999817 | 0.95 |
ENSDART00000137486
|
klhl21
|
kelch-like family member 21 |
| chr7_-_50642150 | 0.95 |
ENSDART00000022918
|
ankrd46b
|
ankyrin repeat domain 46b |
| chr2_-_4277516 | 0.95 |
|
|
|
| chr16_-_19385441 | 0.94 |
ENSDART00000118132
|
5S_rRNA
|
5S ribosomal RNA |
| chr14_-_25154966 | 0.94 |
ENSDART00000148652
|
slc26a2
|
solute carrier family 26 (anion exchanger), member 2 |
| chr10_+_16267331 | 0.93 |
ENSDART00000129844
|
slc12a2
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
| chr6_-_39312306 | 0.93 |
|
|
|
| chr16_-_16614023 | 0.93 |
ENSDART00000160602
|
nbeal2
|
neurobeachin-like 2 |
| chr12_+_23302859 | 0.92 |
ENSDART00000077732
|
bambia
|
BMP and activin membrane-bound inhibitor (Xenopus laevis) homolog a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.4 | GO:0043576 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 1.7 | 5.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 1.2 | 3.5 | GO:0032877 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.5 | 1.5 | GO:0042357 | thiamine metabolic process(GO:0006772) thiamine diphosphate metabolic process(GO:0042357) thiamine-containing compound metabolic process(GO:0042723) |
| 0.5 | 1.8 | GO:0048025 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.4 | 2.0 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.4 | 2.7 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.4 | 1.2 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.4 | 1.5 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.4 | 1.1 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.4 | 4.5 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.4 | 1.5 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.4 | 2.6 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.4 | 1.1 | GO:0015911 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) regulation of organic acid transport(GO:0032890) positive regulation of organic acid transport(GO:0032892) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transport(GO:1903793) regulation of anion transmembrane transport(GO:1903959) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.3 | 2.4 | GO:0006007 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.3 | 1.7 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.3 | 1.7 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.3 | 3.0 | GO:0014896 | regulation of muscle hypertrophy(GO:0014743) muscle hypertrophy(GO:0014896) |
| 0.3 | 1.0 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.3 | 4.8 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.3 | 1.8 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.3 | 5.5 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.3 | 3.0 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.3 | 2.2 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.3 | 1.3 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.2 | 8.0 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.2 | 2.4 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.2 | 1.1 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.2 | 2.1 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.2 | 1.7 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.2 | 2.3 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.2 | 2.2 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.2 | 1.8 | GO:0034333 | adherens junction assembly(GO:0034333) |
| 0.2 | 2.0 | GO:0042026 | protein refolding(GO:0042026) |
| 0.2 | 1.4 | GO:0003207 | cardiac chamber formation(GO:0003207) |
| 0.2 | 1.0 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 | 1.0 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.2 | 2.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.2 | 0.5 | GO:2000391 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.2 | 1.0 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.1 | 2.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.2 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 1.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 1.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 2.2 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 1.2 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 | 2.1 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 2.6 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.4 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) quinolinate metabolic process(GO:0046874) |
| 0.1 | 2.6 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 3.8 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.1 | 0.9 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 0.3 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 0.9 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 1.9 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 2.9 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 0.5 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.1 | 0.9 | GO:0030168 | platelet activation(GO:0030168) |
| 0.1 | 0.4 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 | 0.2 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.1 | 1.6 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 3.0 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 0.9 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 0.4 | GO:0021703 | locus ceruleus development(GO:0021703) |
| 0.1 | 1.8 | GO:0007596 | blood coagulation(GO:0007596) hemostasis(GO:0007599) |
| 0.1 | 4.9 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 0.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 1.5 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.7 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.0 | 2.5 | GO:0048332 | mesoderm morphogenesis(GO:0048332) |
| 0.0 | 1.5 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 1.4 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 1.7 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.0 | 2.2 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.0 | 4.0 | GO:0050770 | regulation of axonogenesis(GO:0050770) |
| 0.0 | 2.4 | GO:0016052 | carbohydrate catabolic process(GO:0016052) |
| 0.0 | 2.0 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.7 | GO:0097581 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.0 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.6 | GO:0071482 | cellular response to light stimulus(GO:0071482) |
| 0.0 | 1.7 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.0 | 0.3 | GO:0045103 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.3 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.3 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 3.4 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 3.2 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 1.4 | GO:0001947 | heart looping(GO:0001947) |
| 0.0 | 0.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.3 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.8 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.3 | 1.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.3 | 2.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.3 | 2.0 | GO:0016234 | inclusion body(GO:0016234) |
| 0.3 | 1.6 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.2 | 2.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 1.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.2 | 8.0 | GO:0030426 | growth cone(GO:0030426) |
| 0.2 | 2.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 2.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.2 | 1.8 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 4.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 1.1 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 1.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 2.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 1.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 2.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 3.4 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 4.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 2.9 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.1 | 1.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.9 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 2.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 3.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 2.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 1.5 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.1 | 4.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 2.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 5.8 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 2.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.7 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 2.1 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.2 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.9 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 4.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.8 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 2.2 | GO:0043005 | neuron projection(GO:0043005) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.4 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 1.7 | 5.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 1.0 | 2.9 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.8 | 7.5 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.7 | 2.2 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.6 | 2.6 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.6 | 3.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.6 | 1.7 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.6 | 5.0 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.4 | 2.2 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.4 | 1.5 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.4 | 2.2 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.3 | 2.4 | GO:0070095 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.3 | 3.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.3 | 1.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.3 | 5.5 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.3 | 2.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.3 | 1.7 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.3 | 1.3 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.2 | 1.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 1.7 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.2 | 3.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.2 | 2.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 2.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 1.5 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.2 | 0.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 1.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 2.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 1.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.7 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 1.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.9 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 0.9 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 1.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.2 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 1.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 2.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 3.7 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.1 | 1.1 | GO:0019203 | carbohydrate phosphatase activity(GO:0019203) sugar-phosphatase activity(GO:0050308) |
| 0.1 | 1.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 2.4 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.3 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 1.8 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 2.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.7 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 1.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 3.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 1.5 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 2.9 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 3.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 4.9 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 4.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.2 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 8.6 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.0 | GO:0004044 | amidophosphoribosyltransferase activity(GO:0004044) |
| 0.0 | 0.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.2 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 4.3 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.5 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 2.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 2.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 3.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 5.1 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.2 | 2.6 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 2.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 2.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 0.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 0.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 2.2 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 1.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.4 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 2.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.8 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |