Project
DANIO-CODE
Navigation
Downloads
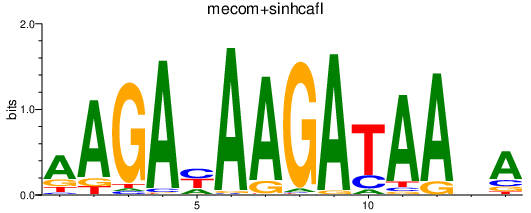
Results for mecom+sinhcafl
Z-value: 2.59
Transcription factors associated with mecom+sinhcafl
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mecom
|
ENSDARG00000060808 | MDS1 and EVI1 complex locus |
|
sinhcafl
|
ENSDARG00000095007 | SIN3-HDAC complex associated factor, like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| fam60al | dr10_dc_chr7_+_28782292_28782387 | -0.58 | 1.9e-02 | Click! |
Activity profile of mecom+sinhcafl motif
Sorted Z-values of mecom+sinhcafl motif
Network of associatons between targets according to the STRING database.
First level regulatory network of mecom+sinhcafl
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_20496283 | 13.99 |
ENSDART00000144047
ENSDART00000104336 |
tnnc2
|
troponin C type 2 (fast) |
| chr17_-_124685 | 10.37 |
ENSDART00000158825
|
actc1b
|
actin, alpha, cardiac muscle 1b |
| chr7_-_24093289 | 7.71 |
ENSDART00000064789
|
txn
|
thioredoxin |
| chr13_+_13550656 | 7.35 |
ENSDART00000057825
|
cfd
|
complement factor D (adipsin) |
| chr15_-_23711689 | 7.11 |
ENSDART00000128644
|
ckmb
|
creatine kinase, muscle b |
| chr16_-_42040346 | 7.05 |
ENSDART00000169313
|
pycard
|
PYD and CARD domain containing |
| chr25_+_18467217 | 6.79 |
ENSDART00000170841
|
cav1
|
caveolin 1 |
| chr14_+_35351559 | 6.61 |
ENSDART00000074685
|
glrbb
|
glycine receptor, beta b |
| chr13_-_33696425 | 6.43 |
ENSDART00000143703
|
flrt3
|
fibronectin leucine rich transmembrane 3 |
| chr15_+_19902697 | 6.38 |
ENSDART00000101204
|
alcamb
|
activated leukocyte cell adhesion molecule b |
| chr23_+_23730717 | 6.15 |
|
|
|
| chr25_-_2485276 | 5.95 |
ENSDART00000154889
ENSDART00000155027 |
BX957346.1
|
ENSDARG00000096850 |
| chr14_-_46651359 | 5.90 |
ENSDART00000163316
|
CR383676.1
|
ENSDARG00000099970 |
| chr21_-_36338514 | 5.57 |
ENSDART00000157826
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr3_+_26896869 | 4.86 |
ENSDART00000065495
|
emp2
|
epithelial membrane protein 2 |
| chr7_-_8128948 | 4.69 |
ENSDART00000057101
|
aep1
|
aerolysin-like protein |
| chr9_+_16437408 | 4.69 |
ENSDART00000006787
|
epha3
|
eph receptor A3 |
| chr21_+_7844259 | 4.66 |
ENSDART00000027268
|
otpa
|
orthopedia homeobox a |
| chr19_+_16318256 | 4.58 |
ENSDART00000137189
|
ptprua
|
protein tyrosine phosphatase, receptor type, U, a |
| chr7_+_38479571 | 4.40 |
ENSDART00000170486
|
f2
|
coagulation factor II (thrombin) |
| chr7_-_16776026 | 4.39 |
ENSDART00000022441
|
dbx1a
|
developing brain homeobox 1a |
| chr10_-_8029671 | 4.23 |
ENSDART00000141445
|
ewsr1a
|
EWS RNA-binding protein 1a |
| chr5_+_49093250 | 4.22 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr23_+_20183765 | 4.19 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr4_+_10722966 | 4.15 |
ENSDART00000102534
ENSDART00000067253 ENSDART00000136000 |
stab2
|
stabilin 2 |
| chr6_-_9964616 | 4.15 |
|
|
|
| chr15_+_46189096 | 4.08 |
|
|
|
| chr2_-_21265803 | 4.05 |
ENSDART00000006870
|
ptgs2a
|
prostaglandin-endoperoxide synthase 2a |
| chr6_-_34024063 | 3.99 |
ENSDART00000003701
|
tie1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr5_+_49093134 | 3.98 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr10_+_22759607 | 3.98 |
|
|
|
| chr8_-_14142049 | 3.91 |
ENSDART00000126432
|
rhoaa
|
ras homolog gene family, member Aa |
| chr11_+_41401893 | 3.89 |
ENSDART00000171655
|
AL929185.1
|
ENSDARG00000103982 |
| chr19_-_35739239 | 3.87 |
|
|
|
| chr22_+_37944019 | 3.86 |
|
|
|
| chr21_-_36338704 | 3.85 |
ENSDART00000157826
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr11_-_22966274 | 3.85 |
ENSDART00000003646
|
optc
|
opticin |
| chr6_-_21894941 | 3.77 |
ENSDART00000160679
|
si:dkey-31g6.6
|
si:dkey-31g6.6 |
| chr20_+_23599157 | 3.76 |
ENSDART00000149922
|
palld
|
palladin, cytoskeletal associated protein |
| chr15_+_42329269 | 3.76 |
ENSDART00000099234
ENSDART00000152731 |
scaf4b
|
SR-related CTD-associated factor 4b |
| chr6_-_7281412 | 3.74 |
ENSDART00000053776
|
fkbp11
|
FK506 binding protein 11 |
| chr25_+_18467835 | 3.70 |
ENSDART00000172338
|
cav1
|
caveolin 1 |
| chr6_+_3519697 | 3.69 |
ENSDART00000013588
|
klhl41b
|
kelch-like family member 41b |
| chr25_-_31218193 | 3.68 |
ENSDART00000170673
ENSDART00000166930 |
lamb1a
|
laminin, beta 1a |
| chr19_+_1814994 | 3.64 |
|
|
|
| chr18_+_16257606 | 3.64 |
ENSDART00000142584
|
alx1
|
ALX homeobox 1 |
| chr14_-_46650724 | 3.56 |
|
|
|
| chr11_-_3275255 | 3.53 |
ENSDART00000066177
|
tuba2
|
tubulin, alpha 2 |
| chr17_+_27706409 | 3.49 |
ENSDART00000123588
ENSDART00000170462 ENSDART00000169708 |
qkia
|
QKI, KH domain containing, RNA binding a |
| chr21_-_15578808 | 3.47 |
ENSDART00000136666
|
mmp11b
|
matrix metallopeptidase 11b |
| chr22_-_3134876 | 3.45 |
ENSDART00000159580
|
lmnb2
|
lamin B2 |
| chr18_-_6494014 | 3.43 |
ENSDART00000062423
|
tnni1c
|
troponin I, skeletal, slow c |
| chr25_+_18467034 | 3.39 |
ENSDART00000170841
|
cav1
|
caveolin 1 |
| chr23_-_24755654 | 3.37 |
ENSDART00000104035
|
ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr6_-_39278328 | 3.36 |
ENSDART00000148531
|
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr3_-_30554400 | 3.34 |
ENSDART00000151097
|
si:ch211-51c14.1
|
si:ch211-51c14.1 |
| chr9_-_34068225 | 3.34 |
ENSDART00000088441
ENSDART00000150035 |
si:ch73-147f11.1
|
si:ch73-147f11.1 |
| chr1_+_21309690 | 3.33 |
ENSDART00000141317
|
dnah6
|
dynein, axonemal, heavy chain 6 |
| chr23_-_32378111 | 3.31 |
ENSDART00000143772
|
dgkaa
|
diacylglycerol kinase, alpha a |
| chr10_+_33462953 | 3.31 |
ENSDART00000137089
|
ryr1a
|
ryanodine receptor 1a (skeletal) |
| chr23_+_9285446 | 3.31 |
ENSDART00000033663
|
rps21
|
ribosomal protein S21 |
| chr1_+_25662910 | 3.29 |
ENSDART00000113020
|
tet2
|
tet methylcytosine dioxygenase 2 |
| chr20_-_34898276 | 3.21 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr10_+_9325719 | 3.16 |
ENSDART00000064968
|
rasgef1bb
|
RasGEF domain family, member 1Bb |
| chr7_+_16615075 | 3.14 |
|
|
|
| chr17_+_53162459 | 3.02 |
|
|
|
| chr20_+_34942798 | 3.02 |
ENSDART00000128895
|
emilin1a
|
elastin microfibril interfacer 1a |
| chr11_-_9479592 | 2.98 |
ENSDART00000175829
|
CU179643.1
|
ENSDARG00000086489 |
| chr24_+_24924379 | 2.96 |
ENSDART00000115165
|
amer2
|
APC membrane recruitment protein 2 |
| chr16_-_42063851 | 2.96 |
ENSDART00000045403
|
etv2
|
ets variant 2 |
| chr22_-_28840467 | 2.92 |
|
|
|
| chr7_+_13753344 | 2.90 |
|
|
|
| chr20_+_26639029 | 2.89 |
ENSDART00000045397
|
stx11b.1
|
syntaxin 11b, tandem duplicate 1 |
| chr3_-_18426055 | 2.87 |
ENSDART00000122968
|
aqp8b
|
aquaporin 8b |
| chr8_+_2428689 | 2.85 |
ENSDART00000081325
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr11_+_18020191 | 2.72 |
ENSDART00000125984
|
mustn1b
|
musculoskeletal, embryonic nuclear protein 1b |
| chr13_+_33378267 | 2.71 |
ENSDART00000025007
|
jag2a
|
jagged 2a |
| chr16_-_14463682 | 2.71 |
ENSDART00000011224
|
itga10
|
integrin, alpha 10 |
| chr1_+_25928481 | 2.70 |
ENSDART00000158193
|
coro2a
|
coronin, actin binding protein, 2A |
| chr14_+_51848088 | 2.67 |
ENSDART00000168874
|
rpl26
|
ribosomal protein L26 |
| chr16_+_46902372 | 2.67 |
ENSDART00000177679
|
thsd7ab
|
thrombospondin, type I, domain containing 7Ab |
| chr7_+_38123920 | 2.63 |
ENSDART00000138669
|
cep89
|
centrosomal protein 89 |
| chr23_-_30114528 | 2.63 |
ENSDART00000131209
|
ccdc187
|
coiled-coil domain containing 187 |
| chr8_-_34077387 | 2.63 |
ENSDART00000159208
ENSDART00000040126 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr14_-_16791447 | 2.60 |
ENSDART00000158002
|
CR812832.1
|
ENSDARG00000103278 |
| chr2_+_29992879 | 2.60 |
ENSDART00000056748
|
en2b
|
engrailed homeobox 2b |
| chr19_+_38033219 | 2.59 |
ENSDART00000158960
|
thsd7aa
|
thrombospondin, type I, domain containing 7Aa |
| chr9_-_19154264 | 2.59 |
ENSDART00000081878
|
pou1f1
|
POU class 1 homeobox 1 |
| chr8_+_3391493 | 2.59 |
ENSDART00000085993
|
pxnb
|
paxillin b |
| chr11_-_13069266 | 2.57 |
ENSDART00000169052
|
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr11_-_3846064 | 2.54 |
ENSDART00000161426
|
gata2a
|
GATA binding protein 2a |
| chr15_+_46141934 | 2.53 |
ENSDART00000154388
|
wu:fb18f06
|
wu:fb18f06 |
| chr23_+_2101659 | 2.48 |
|
|
|
| chr5_+_50379968 | 2.48 |
ENSDART00000050988
|
gcnt4a
|
glucosaminyl (N-acetyl) transferase 4, core 2, a |
| chr8_-_38444845 | 2.45 |
ENSDART00000075989
|
inpp5l
|
inositol polyphosphate-5-phosphatase L |
| chr1_+_19376127 | 2.45 |
ENSDART00000088653
|
prss12
|
protease, serine, 12 (neurotrypsin, motopsin) |
| chr9_-_9370003 | 2.43 |
ENSDART00000132257
|
zgc:113337
|
zgc:113337 |
| chr17_-_30685367 | 2.43 |
ENSDART00000114358
|
zgc:194392
|
zgc:194392 |
| chr8_-_38308535 | 2.41 |
|
|
|
| chr8_+_25060320 | 2.38 |
ENSDART00000000744
|
sypl2b
|
synaptophysin-like 2b |
| chr10_-_44527708 | 2.38 |
|
|
|
| chr13_+_10100146 | 2.34 |
ENSDART00000080805
|
six2a
|
SIX homeobox 2a |
| chr7_-_58751849 | 2.30 |
ENSDART00000165390
|
chmp5b
|
charged multivesicular body protein 5b |
| chr10_-_42157835 | 2.29 |
ENSDART00000141500
ENSDART00000156626 |
BX005309.1
|
ENSDARG00000093535 |
| chr2_-_25325686 | 2.26 |
ENSDART00000111212
|
nck1a
|
NCK adaptor protein 1a |
| chr4_-_19992951 | 2.26 |
ENSDART00000169248
|
drd4-rs
|
dopamine receptor D4 related sequence |
| chr20_+_48971948 | 2.22 |
ENSDART00000164006
|
nkx2.4b
|
NK2 homeobox 4b |
| chr6_-_7281346 | 2.22 |
ENSDART00000133096
|
fkbp11
|
FK506 binding protein 11 |
| chr12_-_7820226 | 2.20 |
ENSDART00000149594
ENSDART00000148866 |
ank3b
|
ankyrin 3b |
| chr3_-_18642744 | 2.18 |
ENSDART00000134208
|
hagh
|
hydroxyacylglutathione hydrolase |
| chr6_+_23611854 | 2.15 |
ENSDART00000167795
|
pik3r6a
|
phosphoinositide-3-kinase, regulatory subunit 6a |
| chr20_-_51746555 | 2.15 |
ENSDART00000065231
|
disp1
|
dispatched homolog 1 (Drosophila) |
| chr18_-_17527321 | 2.13 |
|
|
|
| chr16_+_7745309 | 2.11 |
ENSDART00000081418
ENSDART00000149404 |
bves
|
blood vessel epicardial substance |
| chr14_-_31748733 | 2.07 |
ENSDART00000158014
|
si:ch211-69b22.5
|
si:ch211-69b22.5 |
| chr11_-_9479689 | 2.04 |
ENSDART00000175829
|
CU179643.1
|
ENSDARG00000086489 |
| chr14_-_31522323 | 2.03 |
ENSDART00000172870
|
rbmx
|
RNA binding motif protein, X-linked |
| chr13_+_28382422 | 2.01 |
ENSDART00000043117
|
fbxw4
|
F-box and WD repeat domain containing 4 |
| chr20_+_19175518 | 2.01 |
|
|
|
| chr8_-_31359923 | 1.96 |
ENSDART00000019937
|
gadd45ga
|
growth arrest and DNA-damage-inducible, gamma a |
| chr14_-_31748697 | 1.96 |
ENSDART00000158014
|
si:ch211-69b22.5
|
si:ch211-69b22.5 |
| chr6_-_50205020 | 1.94 |
ENSDART00000083999
ENSDART00000125950 ENSDART00000143050 |
raly
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr22_-_15254208 | 1.92 |
ENSDART00000063008
|
mfng
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr24_-_16993914 | 1.92 |
|
|
|
| chr7_+_9726412 | 1.91 |
ENSDART00000173155
|
adamts17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr22_+_11505448 | 1.91 |
ENSDART00000113930
|
npb
|
neuropeptide B |
| chr8_+_1122000 | 1.91 |
ENSDART00000127252
|
slc35d2
|
solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
| chr2_+_47885551 | 1.91 |
|
|
|
| chr16_-_9075915 | 1.84 |
|
|
|
| chr5_-_41345192 | 1.82 |
|
|
|
| chr3_+_23600462 | 1.81 |
ENSDART00000131410
|
hoxb3a
|
homeobox B3a |
| chr8_+_25060389 | 1.79 |
ENSDART00000000744
|
sypl2b
|
synaptophysin-like 2b |
| chr18_+_45564871 | 1.79 |
|
|
|
| chr24_+_25326286 | 1.74 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| chr20_+_38129564 | 1.74 |
ENSDART00000032161
|
galnt14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr7_+_59806180 | 1.71 |
ENSDART00000131442
|
hspa12b
|
heat shock protein 12B |
| chr8_-_52729505 | 1.66 |
ENSDART00000168241
|
tubb2b
|
tubulin, beta 2b |
| chr8_+_48812632 | 1.62 |
ENSDART00000098363
|
tprg1l
|
tumor protein p63 regulated 1-like |
| chr14_+_33382672 | 1.61 |
ENSDART00000075312
|
apln
|
apelin |
| chr15_-_9616877 | 1.61 |
ENSDART00000168668
|
gab2
|
GRB2-associated binding protein 2 |
| chr4_-_671973 | 1.61 |
ENSDART00000164735
|
rtcb
|
RNA 2',3'-cyclic phosphate and 5'-OH ligase |
| chr3_-_11409342 | 1.59 |
|
|
|
| chr21_-_15578885 | 1.59 |
ENSDART00000136666
|
mmp11b
|
matrix metallopeptidase 11b |
| chr2_-_55673404 | 1.58 |
ENSDART00000158147
|
calr3b
|
calreticulin 3b |
| chr9_+_30909291 | 1.58 |
ENSDART00000146115
|
klf12b
|
Kruppel-like factor 12b |
| chr17_-_12772433 | 1.57 |
ENSDART00000044126
|
ENSDARG00000020655
|
ENSDARG00000020655 |
| chr14_-_33483075 | 1.56 |
ENSDART00000158870
|
si:dkey-76i15.1
|
si:dkey-76i15.1 |
| chr25_+_35796086 | 1.54 |
ENSDART00000112196
|
HIST2H4B
|
zgc:163040 |
| chr12_+_30448812 | 1.54 |
ENSDART00000126064
|
si:ch211-28p3.4
|
si:ch211-28p3.4 |
| chr9_+_30909382 | 1.52 |
ENSDART00000146115
|
klf12b
|
Kruppel-like factor 12b |
| chr6_-_10728582 | 1.52 |
ENSDART00000151102
|
notum2
|
notum pectinacetylesterase 2 |
| chr14_-_2268924 | 1.51 |
ENSDART00000160205
|
pcdh2ab12
|
protocadherin 2 alpha b 12 |
| chr3_-_2010512 | 1.49 |
|
|
|
| chr24_+_25326233 | 1.49 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| chr7_+_44106327 | 1.48 |
ENSDART00000108766
ENSDART00000111441 |
cdh5
|
cadherin 5 |
| chr8_+_27724171 | 1.47 |
ENSDART00000046004
|
wnt2bb
|
wingless-type MMTV integration site family, member 2Bb |
| chr5_-_19691538 | 1.45 |
ENSDART00000139675
|
dao.1
|
D-amino-acid oxidase, tandem duplicate 1 |
| chr2_-_52202004 | 1.43 |
ENSDART00000165350
|
BX908782.1
|
ENSDARG00000098957 |
| chr2_+_26161204 | 1.39 |
ENSDART00000078639
|
slc2a2
|
solute carrier family 2 (facilitated glucose transporter), member 2 |
| chr18_-_39340435 | 1.38 |
ENSDART00000077694
|
leo1
|
LEO1 homolog, Paf1/RNA polymerase II complex component |
| chr11_-_26583383 | 1.37 |
|
|
|
| chr6_-_20625494 | 1.35 |
|
|
|
| chr3_-_22098046 | 1.30 |
ENSDART00000017750
|
myl4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr13_-_11873376 | 1.29 |
ENSDART00000111438
|
mgea5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr1_+_25928586 | 1.29 |
ENSDART00000152643
|
coro2a
|
coronin, actin binding protein, 2A |
| chr13_-_39704878 | 1.29 |
ENSDART00000138981
|
zgc:171482
|
zgc:171482 |
| chr4_+_40119702 | 1.23 |
|
|
|
| chr20_+_33272248 | 1.23 |
ENSDART00000023963
|
ddx1
|
DEAD (Asp-Glu-Ala-Asp) box helicase 1 |
| chr22_-_12905648 | 1.22 |
ENSDART00000157820
|
mfsd6a
|
major facilitator superfamily domain containing 6a |
| chr23_-_29138719 | 1.18 |
ENSDART00000002812
|
casz1
|
castor zinc finger 1 |
| chr6_+_27348529 | 1.16 |
|
|
|
| chr7_-_38340949 | 1.16 |
ENSDART00000173678
|
c1qtnf4
|
C1q and TNF related 4 |
| chr5_-_17542077 | 1.15 |
ENSDART00000144898
|
rnf215
|
ring finger protein 215 |
| chr5_+_22204129 | 1.15 |
|
|
|
| chr2_-_43318809 | 1.13 |
ENSDART00000132588
|
crema
|
cAMP responsive element modulator a |
| chr10_+_22043094 | 1.13 |
ENSDART00000035188
|
kcnip1b
|
Kv channel interacting protein 1 b |
| chr14_-_46650551 | 1.10 |
|
|
|
| chr11_-_26583342 | 1.09 |
|
|
|
| chr15_+_19358079 | 1.09 |
ENSDART00000123815
|
jam3a
|
junctional adhesion molecule 3a |
| chr14_-_10311677 | 1.09 |
ENSDART00000133723
|
si:dkey-92i17.2
|
si:dkey-92i17.2 |
| chr20_-_5324678 | 1.06 |
ENSDART00000114316
|
sptlc2b
|
serine palmitoyltransferase, long chain base subunit 2b |
| chr21_+_21158761 | 1.05 |
ENSDART00000058311
|
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr19_+_17355544 | 1.04 |
ENSDART00000078951
|
MANEAL
|
mannosidase endo-alpha like |
| chr8_+_39478197 | 1.04 |
|
|
|
| chr11_-_41512092 | 1.04 |
ENSDART00000173147
|
FP325130.3
|
ENSDARG00000105426 |
| chr18_+_8388897 | 1.03 |
ENSDART00000083421
|
cpt1b
|
carnitine palmitoyltransferase 1B (muscle) |
| chr15_-_31236277 | 1.02 |
|
|
|
| chr14_-_46650640 | 1.00 |
|
|
|
| KN149797v1_-_70680 | 0.99 |
ENSDART00000170785
|
CABZ01113193.1
|
uncharacterized protein LOC795664 precursor |
| chr10_-_22988906 | 0.98 |
|
|
|
| chr3_-_55278137 | 0.95 |
ENSDART00000123544
|
tex2
|
testis expressed 2 |
| chr17_-_15141309 | 0.94 |
ENSDART00000103405
|
gch1
|
GTP cyclohydrolase 1 |
| chr23_+_37535830 | 0.93 |
|
|
|
| chr5_-_32674787 | 0.93 |
ENSDART00000145222
|
ccbl1
|
cysteine conjugate-beta lyase, cytoplasmic |
| chr6_-_40700033 | 0.91 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr1_+_37033111 | 0.91 |
ENSDART00000139448
|
GALNTL6
|
polypeptide N-acetylgalactosaminyltransferase like 6 |
| chr11_+_38449192 | 0.89 |
ENSDART00000041800
|
epha8
|
eph receptor A8 |
| chr25_+_25343112 | 0.87 |
ENSDART00000161369
|
LRRC56
|
si:ch211-103e16.5 |
| chr7_+_16615154 | 0.85 |
|
|
|
| chr12_-_19160315 | 0.82 |
ENSDART00000152832
|
si:ch211-141o9.10
|
si:ch211-141o9.10 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 13.9 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 1.4 | 4.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 1.3 | 7.7 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 1.2 | 3.6 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 1.2 | 4.7 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 1.1 | 4.4 | GO:0050820 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 1.0 | 4.2 | GO:1901490 | regulation of lymphangiogenesis(GO:1901490) positive regulation of lymphangiogenesis(GO:1901492) |
| 0.9 | 24.9 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.9 | 3.7 | GO:0055016 | hypochord development(GO:0055016) |
| 0.9 | 6.4 | GO:0021885 | forebrain cell migration(GO:0021885) |
| 0.9 | 7.1 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.8 | 2.5 | GO:1902895 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.8 | 3.9 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.8 | 2.3 | GO:0030800 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclic nucleotide metabolic process(GO:0030800) negative regulation of cyclic nucleotide biosynthetic process(GO:0030803) negative regulation of nucleotide biosynthetic process(GO:0030809) negative regulation of cAMP metabolic process(GO:0030815) negative regulation of cAMP biosynthetic process(GO:0030818) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) negative regulation of purine nucleotide biosynthetic process(GO:1900372) |
| 0.7 | 8.2 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) |
| 0.7 | 7.3 | GO:0006956 | complement activation(GO:0006956) |
| 0.7 | 6.6 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.7 | 3.3 | GO:0006211 | 5-methylcytosine catabolic process(GO:0006211) 5-methylcytosine metabolic process(GO:0019857) |
| 0.6 | 4.9 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.6 | 6.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.6 | 3.3 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.5 | 2.7 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.5 | 4.7 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.5 | 1.5 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.5 | 2.0 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.5 | 1.5 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.5 | 1.5 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.4 | 5.8 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.4 | 2.2 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.4 | 4.2 | GO:0000022 | mitotic spindle elongation(GO:0000022) regulation of chondrocyte differentiation(GO:0032330) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.4 | 1.2 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.4 | 3.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.4 | 1.5 | GO:0046416 | D-amino acid catabolic process(GO:0019478) D-serine catabolic process(GO:0036088) D-amino acid metabolic process(GO:0046416) D-serine metabolic process(GO:0070178) |
| 0.4 | 2.1 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.3 | 1.7 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.3 | 3.7 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.3 | 2.6 | GO:0061055 | myotome development(GO:0061055) |
| 0.3 | 6.0 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.3 | 1.4 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) |
| 0.3 | 0.8 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.3 | 3.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.3 | 11.7 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.2 | 2.0 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 0.9 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.2 | 0.6 | GO:0048341 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.2 | 2.5 | GO:0048016 | inositol phosphate-mediated signaling(GO:0048016) |
| 0.2 | 3.3 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.2 | 3.7 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.2 | 0.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.2 | 1.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.2 | 1.9 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.2 | 1.4 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.2 | 5.3 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.2 | 3.0 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.1 | 1.6 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 1.1 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 2.4 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 2.2 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.1 | 5.0 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.1 | 3.2 | GO:1902749 | regulation of cell cycle G2/M phase transition(GO:1902749) |
| 0.1 | 2.6 | GO:0048794 | swim bladder development(GO:0048794) |
| 0.1 | 1.9 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 0.3 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.1 | 0.6 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 3.7 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 1.0 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.1 | 1.1 | GO:0035675 | neuromast hair cell development(GO:0035675) |
| 0.1 | 2.4 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.1 | 0.9 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.7 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 5.2 | GO:0001708 | cell fate specification(GO:0001708) |
| 0.0 | 0.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 2.2 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.0 | 2.7 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 1.6 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 4.6 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 1.2 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.0 | 0.1 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.0 | 2.0 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.4 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 3.6 | GO:0048793 | pronephros development(GO:0048793) |
| 0.0 | 0.7 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.8 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.3 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 4.6 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 4.2 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.7 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 1.7 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 2.6 | GO:0007268 | chemical synaptic transmission(GO:0007268) anterograde trans-synaptic signaling(GO:0098916) |
| 0.0 | 2.0 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 1.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.3 | GO:0048840 | otolith development(GO:0048840) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.0 | GO:0061702 | inflammasome complex(GO:0061702) |
| 1.0 | 21.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.8 | 10.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.6 | 3.2 | GO:0043034 | costamere(GO:0043034) |
| 0.6 | 2.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.6 | 3.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.5 | 4.7 | GO:0046930 | pore complex(GO:0046930) |
| 0.5 | 2.0 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.5 | 16.0 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.3 | 3.7 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.3 | 2.2 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.3 | 3.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.3 | 1.6 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.3 | 3.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.3 | 2.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 0.7 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.2 | 2.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.2 | 0.5 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.2 | 2.2 | GO:0030286 | dynein complex(GO:0030286) |
| 0.2 | 1.4 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.2 | 1.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.7 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 4.9 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 2.7 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 0.5 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 3.7 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 1.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 6.6 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 1.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 2.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 0.6 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 9.0 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 0.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 1.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 25.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 0.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 5.6 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 7.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 6.6 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 2.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.6 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.6 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 3.3 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 2.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.0 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 4.1 | GO:0000139 | Golgi membrane(GO:0000139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 1.4 | 4.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 1.2 | 18.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 1.1 | 6.6 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.9 | 4.7 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.9 | 7.1 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.7 | 3.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.7 | 3.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.6 | 1.9 | GO:0005463 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.6 | 2.3 | GO:0099528 | G-protein coupled neurotransmitter receptor activity(GO:0099528) |
| 0.6 | 3.3 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.5 | 13.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.5 | 1.9 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.5 | 2.9 | GO:0015250 | water channel activity(GO:0015250) |
| 0.4 | 2.5 | GO:0052659 | inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.3 | 4.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.3 | 3.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.3 | 4.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.3 | 7.7 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.3 | 3.4 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.3 | 1.5 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.2 | 0.7 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.2 | 1.6 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.2 | 1.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 1.4 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.2 | 2.1 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.2 | 1.7 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.2 | 4.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 0.8 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.2 | 2.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 14.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.0 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 1.4 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) phosphoprotein binding(GO:0051219) |
| 0.1 | 6.0 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 1.5 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 0.9 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 0.3 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 3.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 5.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 8.2 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 3.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.5 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.8 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 2.1 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 2.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 5.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 2.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 4.0 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 2.2 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.1 | 2.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) amine binding(GO:0043176) serotonin binding(GO:0051378) serotonin receptor activity(GO:0099589) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.0 | 6.4 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 3.6 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.5 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 3.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.8 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 20.7 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 2.9 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 1.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.2 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 1.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 2.4 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.8 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 4.2 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.0 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 13.9 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.6 | 4.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.4 | 8.0 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.3 | 3.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 3.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 2.7 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 1.5 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 1.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 2.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 7.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 2.2 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.0 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.8 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 2.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 14.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 1.8 | 7.3 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 1.4 | 11.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.9 | 4.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.8 | 15.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.6 | 4.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 2.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.3 | 1.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 1.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 0.9 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.9 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 1.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 2.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 3.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 3.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.0 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 2.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 2.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 2.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.5 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.6 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.0 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.1 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |