Project
DANIO-CODE
Navigation
Downloads
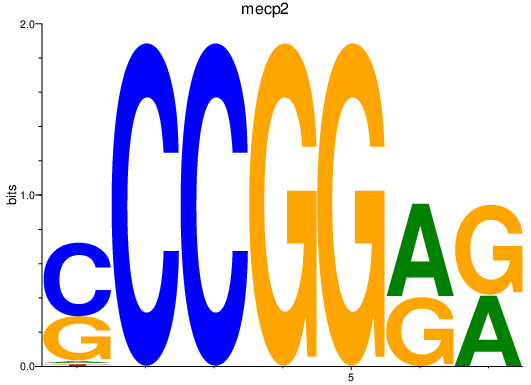
Results for mecp2
Z-value: 1.65
Transcription factors associated with mecp2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mecp2
|
ENSDARG00000014218 | methyl CpG binding protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mecp2 | dr10_dc_chr8_-_7618945_7619005 | -0.01 | 9.6e-01 | Click! |
Activity profile of mecp2 motif
Sorted Z-values of mecp2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of mecp2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| KN149861v1_-_8998 | 2.85 |
|
|
|
| chr1_-_54570813 | 2.15 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr4_-_12931110 | 2.01 |
ENSDART00000013604
|
lemd3
|
LEM domain containing 3 |
| chr25_+_34627031 | 1.94 |
ENSDART00000154377
|
HIST1H2BA (1 of many)
|
histone cluster 1 H2B family member a |
| chr25_+_34549177 | 1.86 |
ENSDART00000157206
|
ENSDARG00000089719
|
ENSDARG00000089719 |
| chr7_-_73628151 | 1.60 |
ENSDART00000123136
|
FP236812.4
|
Histone H2B 1/2 |
| chr19_-_28247275 | 1.56 |
|
|
|
| chr11_+_42265857 | 1.42 |
ENSDART00000039206
|
rps23
|
ribosomal protein S23 |
| chr7_+_73595871 | 1.37 |
ENSDART00000169756
|
HIST1H2BA (1 of many)
|
Histone H2B 1/2 |
| chr18_+_18011562 | 1.34 |
ENSDART00000005027
|
ENSDARG00000011498
|
ENSDARG00000011498 |
| chr4_-_66538843 | 1.34 |
ENSDART00000165173
|
BX548011.1
|
ENSDARG00000103357 |
| chr1_-_54294382 | 1.31 |
ENSDART00000038330
|
khsrp
|
KH-type splicing regulatory protein |
| KN150531v1_-_8098 | 1.28 |
ENSDART00000177246
|
CABZ01080743.1
|
ENSDARG00000106081 |
| chr19_+_43468369 | 1.28 |
ENSDART00000165202
|
pum1
|
pumilio RNA-binding family member 1 |
| chr7_+_5839103 | 1.26 |
ENSDART00000145370
|
zgc:112234
|
zgc:112234 |
| chr17_+_48914379 | 1.25 |
|
|
|
| chr4_-_12930797 | 1.20 |
ENSDART00000108552
|
lemd3
|
LEM domain containing 3 |
| KN149861v1_-_8662 | 1.19 |
ENSDART00000179340
|
CABZ01089907.1
|
ENSDARG00000107716 |
| chr13_+_2345595 | 1.18 |
|
|
|
| chr16_+_11351423 | 1.18 |
ENSDART00000138335
|
gsk3ab
|
glycogen synthase kinase 3 alpha b |
| chr12_-_49212400 | 1.15 |
ENSDART00000112479
|
acadsb
|
acyl-CoA dehydrogenase, short/branched chain |
| chr7_-_10318692 | 1.15 |
ENSDART00000172761
ENSDART00000170476 |
mthfs
|
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chr19_+_14592632 | 1.13 |
ENSDART00000161088
ENSDART00000161965 |
zdhhc18b
|
zinc finger, DHHC-type containing 18b |
| chr20_+_32620786 | 1.12 |
ENSDART00000147319
|
scml4
|
sex comb on midleg-like 4 (Drosophila) |
| chr3_-_7769397 | 1.09 |
|
|
|
| chr5_-_29782745 | 1.08 |
ENSDART00000125381
|
gig2o
|
grass carp reovirus (GCRV)-induced gene 2o |
| chr18_+_25016753 | 1.08 |
ENSDART00000099476
|
fam174b
|
family with sequence similarity 174, member B |
| chr15_-_47307539 | 1.05 |
ENSDART00000027060
|
DMWD
|
DM1 locus, WD repeat containing |
| chr11_-_21203166 | 1.04 |
ENSDART00000080051
|
RASSF5
|
Ras association domain family member 5 |
| chr7_+_41607231 | 1.03 |
ENSDART00000165789
ENSDART00000115090 |
gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr2_-_38305602 | 1.00 |
ENSDART00000061677
|
si:ch211-14a17.6
|
si:ch211-14a17.6 |
| chr1_-_22617455 | 0.99 |
ENSDART00000137567
|
smim14
|
small integral membrane protein 14 |
| chr25_+_4454862 | 0.98 |
ENSDART00000110598
|
zmp:0000001167
|
zmp:0000001167 |
| chr22_-_20141216 | 0.97 |
ENSDART00000128023
|
btbd2a
|
BTB (POZ) domain containing 2a |
| chr4_-_66543241 | 0.95 |
|
|
|
| chr19_+_48144910 | 0.95 |
ENSDART00000149705
|
cdca8
|
cell division cycle associated 8 |
| chr5_-_50972186 | 0.94 |
ENSDART00000134606
|
otpb
|
orthopedia homeobox b |
| chr2_+_10175869 | 0.93 |
ENSDART00000153323
ENSDART00000043499 |
sec22ba
|
SEC22 homolog B, vesicle trafficking protein a |
| chr16_+_33999554 | 0.91 |
ENSDART00000159969
|
arid1aa
|
AT rich interactive domain 1Aa (SWI-like) |
| chr2_+_27835370 | 0.91 |
ENSDART00000078305
|
zswim5
|
zinc finger, SWIM-type containing 5 |
| chr7_-_73590802 | 0.89 |
ENSDART00000167855
|
FP236812.8
|
Histone H2B 1/2 |
| chr3_+_11985816 | 0.86 |
ENSDART00000081367
|
dnaja3a
|
DnaJ (Hsp40) homolog, subfamily A, member 3A |
| chr16_+_33999263 | 0.85 |
ENSDART00000164447
|
arid1aa
|
AT rich interactive domain 1Aa (SWI-like) |
| chr24_-_1014256 | 0.85 |
ENSDART00000114544
|
cdk13
|
cyclin-dependent kinase 13 |
| chr21_+_11683704 | 0.85 |
ENSDART00000081646
|
glrx
|
glutaredoxin (thioltransferase) |
| chr19_+_43468926 | 0.84 |
|
|
|
| chr21_+_11685911 | 0.84 |
ENSDART00000031786
|
glrx
|
glutaredoxin (thioltransferase) |
| chr25_+_34562216 | 0.83 |
ENSDART00000154655
|
CU302436.4
|
ENSDARG00000092743 |
| chr22_-_20141589 | 0.83 |
ENSDART00000085913
|
btbd2a
|
BTB (POZ) domain containing 2a |
| chr22_-_29695242 | 0.82 |
|
|
|
| chr25_-_9889107 | 0.82 |
ENSDART00000137407
|
AL929493.1
|
ENSDARG00000093575 |
| chr12_-_17370920 | 0.81 |
ENSDART00000130735
|
minpp1b
|
multiple inositol-polyphosphate phosphatase 1b |
| chr11_-_27670402 | 0.81 |
ENSDART00000173219
|
eif4g3a
|
eukaryotic translation initiation factor 4 gamma, 3a |
| chr25_-_13307171 | 0.81 |
ENSDART00000056723
|
gins3
|
GINS complex subunit 3 |
| chr15_+_38397772 | 0.81 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr22_+_132285 | 0.80 |
ENSDART00000059140
|
slc25a20
|
solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 |
| chr21_+_4091665 | 0.77 |
ENSDART00000148138
|
lrrc8aa
|
leucine rich repeat containing 8 family, member Aa |
| chr8_-_22265875 | 0.76 |
ENSDART00000134033
|
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr16_+_42567707 | 0.75 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr7_-_41601001 | 0.75 |
ENSDART00000174258
|
zgc:92818
|
zgc:92818 |
| chr5_-_40503137 | 0.74 |
ENSDART00000051070
|
golph3
|
golgi phosphoprotein 3 |
| chr24_+_12689711 | 0.74 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr16_+_33999719 | 0.73 |
ENSDART00000159969
|
arid1aa
|
AT rich interactive domain 1Aa (SWI-like) |
| chr11_-_21203114 | 0.71 |
ENSDART00000080051
|
RASSF5
|
Ras association domain family member 5 |
| chr1_-_53021646 | 0.71 |
|
|
|
| chr12_-_13692190 | 0.70 |
ENSDART00000152370
|
foxh1
|
forkhead box H1 |
| chr21_+_11686037 | 0.70 |
ENSDART00000031786
|
glrx
|
glutaredoxin (thioltransferase) |
| chr8_-_4270732 | 0.69 |
ENSDART00000134378
|
cux2b
|
cut-like homeobox 2b |
| chr19_+_14490203 | 0.69 |
ENSDART00000164386
|
arid1ab
|
AT rich interactive domain 1Ab (SWI-like) |
| chr19_-_3785774 | 0.69 |
ENSDART00000161738
|
smim13
|
small integral membrane protein 13 |
| chr11_-_44756789 | 0.69 |
ENSDART00000161712
|
syngr2b
|
synaptogyrin 2b |
| chr14_-_47210912 | 0.68 |
ENSDART00000031640
ENSDART00000132966 |
zgc:113425
|
zgc:113425 |
| chr15_-_30935719 | 0.68 |
ENSDART00000050649
|
msi2b
|
musashi RNA-binding protein 2b |
| chr10_-_25383262 | 0.68 |
ENSDART00000166348
|
cct8
|
chaperonin containing TCP1, subunit 8 (theta) |
| chr24_+_12689887 | 0.68 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr23_-_27768575 | 0.67 |
ENSDART00000146703
|
IKZF4
|
IKAROS family zinc finger 4 |
| chr21_+_13136338 | 0.67 |
ENSDART00000142569
|
specc1lb
|
sperm antigen with calponin homology and coiled-coil domains 1-like b |
| chr19_+_14490375 | 0.67 |
ENSDART00000164386
|
arid1ab
|
AT rich interactive domain 1Ab (SWI-like) |
| chr5_-_8712114 | 0.66 |
ENSDART00000134896
ENSDART00000147000 |
lrp13
|
low-density lipoprotein receptor related-protein 13 |
| chr21_+_45622590 | 0.66 |
ENSDART00000162422
|
sar1b
|
secretion associated, Ras related GTPase 1B |
| chr7_+_31750514 | 0.66 |
ENSDART00000173976
|
mettl15
|
methyltransferase like 15 |
| chr2_+_39142628 | 0.66 |
ENSDART00000179419
|
clstn2
|
calsyntenin 2 |
| chr15_-_34700453 | 0.65 |
ENSDART00000140882
|
ispd
|
isoprenoid synthase domain containing |
| chr21_+_11686197 | 0.64 |
ENSDART00000031786
|
glrx
|
glutaredoxin (thioltransferase) |
| chr16_-_42116471 | 0.63 |
ENSDART00000058620
|
zp3d.1
|
zona pellucida glycoprotein 3d tandem duplicate 1 |
| chr7_+_15479700 | 0.63 |
|
|
|
| KN149861v1_-_6916 | 0.62 |
ENSDART00000169003
|
5_8S_rRNA
|
5.8S ribosomal RNA |
| chr16_-_23885481 | 0.62 |
ENSDART00000139964
|
rps27.2
|
ribosomal protein S27, isoform 2 |
| chr5_-_13145749 | 0.61 |
ENSDART00000166957
|
purba
|
purine-rich element binding protein Ba |
| chr23_+_540664 | 0.61 |
ENSDART00000034707
|
lsm14b
|
LSM family member 14B |
| chr23_+_41015697 | 0.61 |
ENSDART00000115161
|
reps2
|
RALBP1 associated Eps domain containing 2 |
| chr17_-_49910160 | 0.60 |
ENSDART00000122747
|
tmem30aa
|
transmembrane protein 30Aa |
| chr19_-_32313943 | 0.59 |
ENSDART00000113797
|
zbtb10
|
zinc finger and BTB domain containing 10 |
| chr7_-_10318801 | 0.59 |
ENSDART00000172761
ENSDART00000170476 |
mthfs
|
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chr5_+_65491173 | 0.59 |
ENSDART00000050847
ENSDART00000172117 |
GLDC
|
glycine decarboxylase |
| chr19_+_14132374 | 0.59 |
ENSDART00000164696
|
tmem222b
|
transmembrane protein 222b |
| chr19_-_42892975 | 0.58 |
ENSDART00000131715
|
psmb4
|
proteasome subunit beta 4 |
| KN149861v1_-_8062 | 0.58 |
ENSDART00000179340
|
CABZ01089907.1
|
ENSDARG00000107716 |
| chr14_-_44880144 | 0.58 |
ENSDART00000163543
|
abhd18
|
abhydrolase domain containing 18 |
| chr10_-_35488523 | 0.57 |
|
|
|
| chr17_-_23874694 | 0.57 |
ENSDART00000122108
|
pdzd8
|
PDZ domain containing 8 |
| chr3_+_27476229 | 0.57 |
ENSDART00000024453
|
usp7
|
ubiquitin specific peptidase 7 (herpes virus-associated) |
| chr7_+_16096816 | 0.57 |
ENSDART00000002449
|
mpped2a
|
metallophosphoesterase domain containing 2a |
| chr1_-_54294205 | 0.57 |
ENSDART00000140016
|
khsrp
|
KH-type splicing regulatory protein |
| chr19_+_14592518 | 0.56 |
ENSDART00000161088
ENSDART00000161965 |
zdhhc18b
|
zinc finger, DHHC-type containing 18b |
| chr10_-_2655016 | 0.56 |
|
|
|
| KN149883v1_-_5164 | 0.56 |
ENSDART00000158506
ENSDART00000172360 |
NDUFA13
|
NADH:ubiquinone oxidoreductase subunit A13 |
| chr22_-_3165441 | 0.56 |
ENSDART00000158009
|
lonp1
|
lon peptidase 1, mitochondrial |
| chr8_-_22265802 | 0.56 |
ENSDART00000134033
|
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr16_+_22949379 | 0.55 |
ENSDART00000137783
|
flad1
|
flavin adenine dinucleotide synthetase 1 |
| chr22_+_26736632 | 0.55 |
|
|
|
| chr23_-_24328931 | 0.55 |
|
|
|
| chr19_+_770458 | 0.55 |
ENSDART00000062518
|
gstr
|
glutathione S-transferase rho |
| chr22_-_5722123 | 0.55 |
|
|
|
| chr24_-_37810565 | 0.55 |
ENSDART00000172178
|
si:ch211-231f6.6
|
si:ch211-231f6.6 |
| chr1_+_14572155 | 0.55 |
ENSDART00000033018
|
pi4k2b
|
phosphatidylinositol 4-kinase type 2 beta |
| chr14_+_300285 | 0.55 |
ENSDART00000159949
|
wdr1
|
WD repeat domain 1 |
| chr17_-_23874858 | 0.54 |
ENSDART00000122108
|
pdzd8
|
PDZ domain containing 8 |
| chr14_+_42109615 | 0.54 |
|
|
|
| chr7_-_28771601 | 0.53 |
ENSDART00000052342
|
thap11
|
THAP domain containing 11 |
| chr2_+_50327729 | 0.53 |
ENSDART00000097939
|
zcchc2
|
zinc finger, CCHC domain containing 2 |
| chr19_+_41894734 | 0.52 |
ENSDART00000087187
|
ago2
|
argonaute RISC catalytic component 2 |
| chr3_-_10615725 | 0.52 |
ENSDART00000048095
ENSDART00000155152 |
elac2
|
elaC ribonuclease Z 2 |
| chr24_-_1014318 | 0.52 |
ENSDART00000114544
|
cdk13
|
cyclin-dependent kinase 13 |
| chr19_+_44185325 | 0.52 |
ENSDART00000063870
|
rpl11
|
ribosomal protein L11 |
| chr16_+_22949494 | 0.52 |
ENSDART00000137783
|
flad1
|
flavin adenine dinucleotide synthetase 1 |
| chr2_+_38305772 | 0.52 |
ENSDART00000170672
|
nedd8l
|
neural precursor cell expressed, developmentally down-regulated 8, like |
| chr5_-_40503363 | 0.52 |
ENSDART00000074781
|
golph3
|
golgi phosphoprotein 3 |
| chr25_-_19388415 | 0.52 |
ENSDART00000156016
|
zgc:193812
|
zgc:193812 |
| chr23_+_12426885 | 0.51 |
ENSDART00000143728
|
pigt
|
phosphatidylinositol glycan anchor biosynthesis, class T |
| chr19_+_41894906 | 0.51 |
ENSDART00000087187
|
ago2
|
argonaute RISC catalytic component 2 |
| chr24_-_23973276 | 0.51 |
ENSDART00000111096
|
zgc:112982
|
zgc:112982 |
| KN149861v1_-_6089 | 0.51 |
|
|
|
| chr8_-_22266041 | 0.50 |
ENSDART00000134033
|
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr20_+_54595334 | 0.50 |
|
|
|
| chr19_+_1298795 | 0.49 |
ENSDART00000113368
|
RNF5
|
ring finger protein 5 |
| chr24_+_24140538 | 0.49 |
ENSDART00000122294
|
ythdf3
|
YTH N(6)-methyladenosine RNA binding protein 3 |
| chr20_-_30132364 | 0.49 |
ENSDART00000033588
|
BX248324.1
|
ENSDARG00000024870 |
| chr9_-_32347822 | 0.49 |
ENSDART00000037182
|
ankrd44
|
ankyrin repeat domain 44 |
| chr21_-_25576567 | 0.48 |
ENSDART00000115276
ENSDART00000137896 |
fibpb
|
fibroblast growth factor (acidic) intracellular binding protein b |
| chr1_-_51870424 | 0.48 |
ENSDART00000135636
|
acy3.1
|
aspartoacylase (aminocyclase) 3, tandem duplicate 1 |
| chr21_-_44707514 | 0.48 |
ENSDART00000013814
|
ammecr1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 homolog (human) |
| chr6_+_6640324 | 0.47 |
ENSDART00000150967
|
si:ch211-85n16.3
|
si:ch211-85n16.3 |
| chr8_+_388003 | 0.47 |
ENSDART00000067668
|
pym1
|
PYM homolog 1, exon junction complex associated factor |
| chr14_-_6096258 | 0.47 |
|
|
|
| chr17_-_6225136 | 0.46 |
ENSDART00000137389
ENSDART00000115389 |
txlnbb
|
taxilin beta b |
| chr10_+_3427790 | 0.46 |
ENSDART00000081599
|
ptpn11a
|
protein tyrosine phosphatase, non-receptor type 11, a |
| chr10_+_43208682 | 0.46 |
ENSDART00000160159
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr20_+_32620937 | 0.46 |
ENSDART00000147319
|
scml4
|
sex comb on midleg-like 4 (Drosophila) |
| chr23_-_27579257 | 0.46 |
ENSDART00000137229
|
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr19_+_14592246 | 0.46 |
ENSDART00000161088
ENSDART00000161965 |
zdhhc18b
|
zinc finger, DHHC-type containing 18b |
| chr2_-_9946238 | 0.45 |
ENSDART00000097732
|
dvl3a
|
dishevelled segment polarity protein 3a |
| chr7_-_51497945 | 0.45 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr11_-_25615491 | 0.45 |
ENSDART00000145655
|
tmem51b
|
transmembrane protein 51b |
| chr18_+_5303006 | 0.45 |
|
|
|
| chr2_-_9946148 | 0.45 |
ENSDART00000175460
|
dvl3a
|
dishevelled segment polarity protein 3a |
| chr23_+_20505319 | 0.45 |
ENSDART00000140219
|
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr9_-_23431733 | 0.45 |
ENSDART00000053282
|
ccnt2a
|
cyclin T2a |
| chr17_-_27217309 | 0.45 |
ENSDART00000130080
|
asap3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
| chr7_-_26260802 | 0.44 |
ENSDART00000121698
|
senp3b
|
SUMO1/sentrin/SMT3 specific peptidase 3b |
| chr5_+_22173976 | 0.44 |
ENSDART00000142112
|
vma21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr13_+_13875110 | 0.44 |
ENSDART00000131875
|
atrn
|
attractin |
| KN149861v1_-_6661 | 0.44 |
|
|
|
| chr16_+_17762256 | 0.44 |
ENSDART00000128672
|
tmem238b
|
transmembrane protein 238b |
| chr19_-_3785709 | 0.44 |
ENSDART00000165947
|
CT030005.1
|
ENSDARG00000099249 |
| chr22_+_11114323 | 0.44 |
ENSDART00000047442
|
bcor
|
BCL6 corepressor |
| chr12_-_29928951 | 0.43 |
ENSDART00000149077
ENSDART00000148847 |
trub1
|
TruB pseudouridine (psi) synthase family member 1 |
| chr8_-_4517590 | 0.43 |
ENSDART00000090731
|
dhx37
|
DEAH (Asp-Glu-Ala-His) box polypeptide 37 |
| chr21_+_8334453 | 0.43 |
ENSDART00000055336
|
dennd1a
|
DENN/MADD domain containing 1A |
| chr2_+_37855019 | 0.43 |
ENSDART00000113337
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr11_+_40847576 | 0.43 |
ENSDART00000041531
|
park7
|
parkinson protein 7 |
| chr20_-_45757288 | 0.43 |
ENSDART00000124582
ENSDART00000100290 |
napbb
|
N-ethylmaleimide-sensitive factor attachment protein, beta b |
| chr18_-_46371631 | 0.43 |
ENSDART00000018163
|
irf2bp1
|
interferon regulatory factor 2 binding protein 1 |
| chr21_+_11685992 | 0.42 |
ENSDART00000031786
|
glrx
|
glutaredoxin (thioltransferase) |
| chr23_-_5188577 | 0.42 |
ENSDART00000123191
|
ube2t
|
ubiquitin-conjugating enzyme E2T (putative) |
| chr18_+_19893842 | 0.42 |
ENSDART00000165336
|
zgc:162898
|
zgc:162898 |
| chr19_-_48144752 | 0.42 |
ENSDART00000158979
|
c19h1orf109
|
c19h1orf109 homolog (H. sapiens) |
| chr24_+_5176362 | 0.42 |
ENSDART00000155926
|
si:ch73-206p6.1
|
si:ch73-206p6.1 |
| chr20_+_32620873 | 0.42 |
ENSDART00000147319
|
scml4
|
sex comb on midleg-like 4 (Drosophila) |
| chr7_-_30654776 | 0.42 |
ENSDART00000075421
|
sord
|
sorbitol dehydrogenase |
| chr4_+_9910588 | 0.41 |
ENSDART00000159762
|
sbf1
|
SET binding factor 1 |
| chr17_+_52438099 | 0.41 |
ENSDART00000135246
|
dlst
|
dihydrolipoamide S-succinyltransferase |
| chr1_-_50626230 | 0.41 |
|
|
|
| chr19_+_41895061 | 0.40 |
ENSDART00000087187
|
ago2
|
argonaute RISC catalytic component 2 |
| chr23_+_540624 | 0.40 |
ENSDART00000034707
|
lsm14b
|
LSM family member 14B |
| chr21_-_13026036 | 0.40 |
ENSDART00000135623
|
fam219aa
|
family with sequence similarity 219, member Aa |
| KN150307v1_+_6450 | 0.40 |
ENSDART00000159959
|
CABZ01038161.1
|
ENSDARG00000098784 |
| chr19_+_3115685 | 0.40 |
ENSDART00000127473
ENSDART00000126549 ENSDART00000024593 ENSDART00000082353 ENSDART00000141324 |
hsf1
|
heat shock transcription factor 1 |
| chr2_+_38305689 | 0.40 |
ENSDART00000170672
|
nedd8l
|
neural precursor cell expressed, developmentally down-regulated 8, like |
| chr2_-_37418967 | 0.40 |
ENSDART00000015723
|
prkci
|
protein kinase C, iota |
| chr20_-_9440244 | 0.39 |
ENSDART00000025330
|
rdh14b
|
retinol dehydrogenase 14b (all-trans/9-cis/11-cis) |
| chr9_+_331815 | 0.39 |
ENSDART00000170312
|
actr5
|
ARP5 actin related protein 5 homolog |
| chr7_-_6308104 | 0.39 |
ENSDART00000173010
|
zgc:112234
|
zgc:112234 |
| chr19_-_48144715 | 0.39 |
ENSDART00000158979
|
c19h1orf109
|
c19h1orf109 homolog (H. sapiens) |
| chr2_-_47766563 | 0.39 |
ENSDART00000038228
|
ap1s3b
|
adaptor-related protein complex 1, sigma 3 subunit, b |
| chr25_-_29914405 | 0.39 |
ENSDART00000171137
|
pdia3
|
protein disulfide isomerase family A, member 3 |
| chr9_-_32532843 | 0.39 |
ENSDART00000078499
|
rftn2
|
raftlin family member 2 |
| chr25_-_13562597 | 0.39 |
ENSDART00000045488
|
csnk2a2b
|
casein kinase 2, alpha prime polypeptide b |
| chr25_+_35793115 | 0.39 |
ENSDART00000103007
|
zgc:173552
|
zgc:173552 |
| chr20_-_48255596 | 0.38 |
ENSDART00000031167
|
tfap2d
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.8 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.4 | 1.1 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.4 | 2.1 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.3 | 1.4 | GO:0072387 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.2 | 3.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.2 | 1.7 | GO:0035999 | folic acid-containing compound biosynthetic process(GO:0009396) tetrahydrofolate interconversion(GO:0035999) |
| 0.2 | 0.5 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.2 | 0.7 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.2 | 0.7 | GO:0070861 | regulation of COPII vesicle coating(GO:0003400) regulation of protein exit from endoplasmic reticulum(GO:0070861) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.2 | 0.5 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.2 | 0.6 | GO:0030728 | ovulation(GO:0030728) |
| 0.2 | 0.5 | GO:1903259 | exon-exon junction complex disassembly(GO:1903259) |
| 0.2 | 0.5 | GO:0042698 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) |
| 0.1 | 0.4 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.4 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.1 | 0.7 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.1 | 0.5 | GO:0071867 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.1 | 0.9 | GO:0090177 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.4 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.1 | 0.4 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 1.0 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.4 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 1.3 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.1 | 0.3 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.1 | 0.7 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.1 | 0.4 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.1 | 0.5 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 1.6 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.4 | GO:0046078 | dUMP biosynthetic process(GO:0006226) dUMP metabolic process(GO:0046078) |
| 0.1 | 0.4 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.1 | 2.2 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.1 | 2.1 | GO:0019827 | stem cell population maintenance(GO:0019827) |
| 0.1 | 0.3 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 0.5 | GO:0050427 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 | 0.2 | GO:2000737 | negative regulation of stem cell differentiation(GO:2000737) |
| 0.1 | 0.3 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 0.2 | GO:1903010 | regulation of bone development(GO:1903010) |
| 0.1 | 1.1 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.9 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.1 | 3.7 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.1 | 0.6 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 0.2 | GO:0043420 | anthranilate metabolic process(GO:0043420) |
| 0.1 | 0.4 | GO:0019405 | alditol catabolic process(GO:0019405) |
| 0.1 | 0.4 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.8 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 1.7 | GO:0009408 | response to heat(GO:0009408) |
| 0.1 | 0.2 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.1 | 1.7 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 1.2 | GO:0097192 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.0 | 0.6 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.5 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.0 | 0.4 | GO:0032447 | protein urmylation(GO:0032447) |
| 0.0 | 0.5 | GO:0061157 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.0 | 0.6 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.5 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) estrogen metabolic process(GO:0008210) |
| 0.0 | 0.4 | GO:0051351 | positive regulation of ligase activity(GO:0051351) |
| 0.0 | 0.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.5 | GO:0034033 | nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 | 0.1 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.5 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.2 | GO:1990709 | presynaptic active zone organization(GO:1990709) |
| 0.0 | 0.4 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.0 | 0.4 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.6 | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032435) negative regulation of proteasomal protein catabolic process(GO:1901799) negative regulation of proteolysis involved in cellular protein catabolic process(GO:1903051) |
| 0.0 | 0.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.3 | GO:0030534 | adult behavior(GO:0030534) |
| 0.0 | 0.3 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.4 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.0 | 0.2 | GO:1902315 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.0 | 1.6 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.1 | GO:1902895 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.0 | 0.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.1 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.2 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.2 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.0 | 1.1 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.1 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) ribonucleoprotein complex disassembly(GO:0032988) |
| 0.0 | 0.9 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.1 | GO:0003190 | heart valve formation(GO:0003188) atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.4 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.1 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 0.3 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.4 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.9 | GO:1902667 | regulation of axon guidance(GO:1902667) |
| 0.0 | 0.5 | GO:0060119 | inner ear receptor cell development(GO:0060119) |
| 0.0 | 0.3 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.4 | GO:0032456 | endocytic recycling(GO:0032456) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.3 | GO:0035060 | brahma complex(GO:0035060) |
| 0.3 | 3.2 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.2 | 1.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.2 | 2.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.2 | 0.9 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.2 | 2.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.2 | 0.8 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.9 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 0.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.5 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 0.4 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.6 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 2.1 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 0.8 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 0.3 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.1 | 0.7 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 0.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.1 | 1.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 2.0 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 0.4 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.3 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 2.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.7 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.2 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 0.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 2.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.2 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.0 | 0.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.7 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.4 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.9 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.5 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.4 | 2.1 | GO:0070551 | endoribonuclease activity, cleaving siRNA-paired mRNA(GO:0070551) |
| 0.4 | 1.2 | GO:0034417 | bisphosphoglycerate 3-phosphatase activity(GO:0034417) |
| 0.3 | 1.4 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.2 | 1.9 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 0.6 | GO:0071424 | rRNA (cytosine-N4-)-methyltransferase activity(GO:0071424) |
| 0.2 | 0.5 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.2 | 0.5 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.2 | 0.5 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 0.1 | 3.6 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 0.8 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.5 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 1.4 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.5 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 1.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.4 | GO:0070260 | 5'-tyrosyl-DNA phosphodiesterase activity(GO:0070260) |
| 0.1 | 0.5 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 1.6 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.8 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.5 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.5 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 1.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.5 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 1.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.4 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.6 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.5 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.2 | GO:0004352 | glutamate dehydrogenase (NAD+) activity(GO:0004352) glutamate dehydrogenase [NAD(P)+] activity(GO:0004353) oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 2.6 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 2.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 3.3 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.2 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.5 | GO:0034595 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.0 | 0.5 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.3 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.2 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.3 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.8 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 1.4 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.2 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 1.1 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.4 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.5 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.1 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.0 | 0.5 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 1.2 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 0.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.4 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 0.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 2.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.5 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.5 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.5 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 2.9 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.5 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.8 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 1.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 0.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 0.8 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.5 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.0 | 1.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.2 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.7 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 0.1 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.9 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.5 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |