Project
DANIO-CODE
Navigation
Downloads
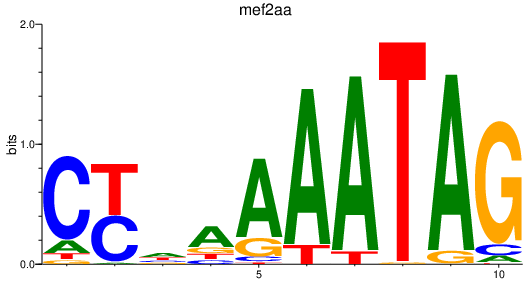
Results for mef2aa
Z-value: 2.55
Transcription factors associated with mef2aa
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mef2aa
|
ENSDARG00000031756 | myocyte enhancer factor 2aa |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mef2aa | dr10_dc_chr18_+_23198330_23198352 | 0.96 | 2.0e-09 | Click! |
Activity profile of mef2aa motif
Sorted Z-values of mef2aa motif
Network of associatons between targets according to the STRING database.
First level regulatory network of mef2aa
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_60973097 | 10.23 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr17_-_124685 | 9.64 |
ENSDART00000158825
|
actc1b
|
actin, alpha, cardiac muscle 1b |
| chr1_-_12584282 | 9.22 |
ENSDART00000127838
|
pcdh18a
|
protocadherin 18a |
| chr3_-_32686790 | 8.67 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr7_+_6814828 | 8.64 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr12_-_17590507 | 8.04 |
ENSDART00000010144
|
pvalb2
|
parvalbumin 2 |
| chr19_+_2162466 | 7.44 |
|
|
|
| chr12_-_5085227 | 7.05 |
ENSDART00000160729
|
rbp4
|
retinol binding protein 4, plasma |
| chr19_-_7531709 | 6.98 |
ENSDART00000104750
|
mllt11
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 11 |
| chr1_-_12584179 | 6.86 |
|
|
|
| chr1_+_6862901 | 6.86 |
ENSDART00000015732
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr2_-_28446615 | 6.82 |
ENSDART00000179495
|
cdh6
|
cadherin 6 |
| chr5_-_71340996 | 6.82 |
ENSDART00000162526
|
smyd1a
|
SET and MYND domain containing 1a |
| chr3_+_28808901 | 6.28 |
ENSDART00000141904
ENSDART00000077221 |
lgals1l1
|
lectin, galactoside-binding, soluble, 1 (galectin 1)-like 1 |
| chr5_+_38236956 | 5.82 |
ENSDART00000160236
|
fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr5_-_28090077 | 5.74 |
|
|
|
| chr1_-_50066633 | 5.41 |
|
|
|
| chr15_+_7068253 | 5.40 |
ENSDART00000114560
|
pik3cb
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
| chr5_+_66712197 | 5.38 |
ENSDART00000014822
|
ebf2
|
early B-cell factor 2 |
| chr12_+_25684420 | 5.33 |
ENSDART00000024415
|
epas1a
|
endothelial PAS domain protein 1a |
| chr8_-_11191783 | 5.31 |
ENSDART00000131171
|
unc45b
|
unc-45 myosin chaperone B |
| chr1_+_16683931 | 5.29 |
ENSDART00000103262
ENSDART00000145068 ENSDART00000169619 ENSDART00000010526 |
fat1a
|
FAT atypical cadherin 1a |
| chr23_+_20183765 | 5.29 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr12_-_25973094 | 5.24 |
ENSDART00000171206
ENSDART00000171212 ENSDART00000170265 |
ldb3b
|
LIM domain binding 3b |
| chr16_+_5466342 | 5.24 |
ENSDART00000160008
|
plecb
|
plectin b |
| chr12_+_6007990 | 5.21 |
ENSDART00000091868
|
g6pca.2
|
glucose-6-phosphatase a, catalytic subunit, tandem duplicate 2 |
| chr15_+_24064257 | 5.15 |
ENSDART00000156714
|
CR394545.1
|
ENSDARG00000097097 |
| chr22_-_6977038 | 5.07 |
ENSDART00000133143
ENSDART00000146813 |
gpd1b
|
glycerol-3-phosphate dehydrogenase 1b |
| chr14_+_21816442 | 4.98 |
ENSDART00000109759
|
tmx2a
|
thioredoxin-related transmembrane protein 2a |
| chr7_-_22685672 | 4.97 |
ENSDART00000101447
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr1_-_10391257 | 4.92 |
ENSDART00000102903
|
dmd
|
dystrophin |
| chr8_-_17480730 | 4.78 |
ENSDART00000100667
|
skia
|
v-ski avian sarcoma viral oncogene homolog a |
| chr18_-_26483645 | 4.66 |
|
|
|
| chr3_+_34064081 | 4.55 |
|
|
|
| chr10_-_8074713 | 4.50 |
ENSDART00000140476
|
atp6v0a2a
|
ATPase, H+ transporting, lysosomal V0 subunit a2a |
| chr16_-_29229687 | 4.50 |
ENSDART00000132589
|
mef2d
|
myocyte enhancer factor 2d |
| chr20_+_15653121 | 4.41 |
ENSDART00000152702
|
jun
|
jun proto-oncogene |
| chr1_-_30772653 | 4.37 |
ENSDART00000087115
|
rims1b
|
regulating synaptic membrane exocytosis 1b |
| chr3_-_39031305 | 4.24 |
ENSDART00000022393
|
si:dkeyp-57f11.2
|
si:dkeyp-57f11.2 |
| chr24_+_26922969 | 4.22 |
|
|
|
| chr8_+_33010788 | 4.04 |
ENSDART00000172672
|
angptl2b
|
angiopoietin-like 2b |
| chr8_+_10267246 | 3.99 |
ENSDART00000159312
ENSDART00000160766 |
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr7_+_59344089 | 3.91 |
ENSDART00000146384
|
rpl34
|
ribosomal protein L34 |
| chr15_+_19717045 | 3.83 |
ENSDART00000138680
|
lim2.3
|
lens intrinsic membrane protein 2.3 |
| chr7_-_23474701 | 3.79 |
ENSDART00000048050
|
ITGB1BP2
|
integrin subunit beta 1 binding protein 2 |
| chr15_-_27778111 | 3.74 |
ENSDART00000134373
|
lhx1a
|
LIM homeobox 1a |
| chr2_+_22839514 | 3.71 |
ENSDART00000167915
|
lrrc8da
|
leucine rich repeat containing 8 family, member Da |
| KN150214v1_-_155944 | 3.71 |
|
|
|
| chr15_+_7068009 | 3.70 |
ENSDART00000155268
|
pik3cb
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
| chr24_+_19842162 | 3.69 |
ENSDART00000123031
|
CR381686.3
|
ENSDARG00000094073 |
| chr14_-_29542922 | 3.68 |
|
|
|
| chr1_+_51706536 | 3.61 |
|
|
|
| chr17_-_14663139 | 3.60 |
ENSDART00000037371
|
ppp1r13ba
|
protein phosphatase 1, regulatory subunit 13Ba |
| chr23_+_17856053 | 3.58 |
ENSDART00000154427
|
CR381647.1
|
ENSDARG00000097211 |
| chr13_+_24703802 | 3.57 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr16_-_31517906 | 3.57 |
ENSDART00000145691
|
ENSDARG00000054814
|
ENSDARG00000054814 |
| chr23_-_39956151 | 3.52 |
ENSDART00000115330
|
ppp1r14c
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr10_+_36710393 | 3.47 |
|
|
|
| chr13_+_22350043 | 3.42 |
ENSDART00000136863
|
ldb3a
|
LIM domain binding 3a |
| chr17_-_14551031 | 3.41 |
ENSDART00000162452
|
daam1a
|
dishevelled associated activator of morphogenesis 1a |
| chr15_+_9351511 | 3.32 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr25_-_22821132 | 3.30 |
|
|
|
| chr14_-_26999410 | 3.25 |
ENSDART00000159727
|
pcdh11
|
protocadherin 11 |
| chr18_+_26737365 | 3.19 |
ENSDART00000141672
|
alpk3a
|
alpha-kinase 3a |
| chr24_-_25545773 | 3.18 |
ENSDART00000015391
|
chrnd
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr1_-_5796394 | 3.14 |
ENSDART00000109356
|
klf7a
|
Kruppel-like factor 7a |
| chr19_+_25892125 | 3.12 |
ENSDART00000112808
|
col28a1a
|
collagen, type XXVIII, alpha 1a |
| chr5_+_23802054 | 3.09 |
ENSDART00000144226
|
ctsll
|
cathepsin L, like |
| chr18_+_23204736 | 3.05 |
ENSDART00000171594
ENSDART00000148106 |
mef2aa
|
myocyte enhancer factor 2aa |
| chr10_-_22875997 | 3.04 |
ENSDART00000079454
|
vamp2
|
vesicle-associated membrane protein 2 |
| chr5_+_69036145 | 3.00 |
ENSDART00000174403
|
fsd1l
|
fibronectin type III and SPRY domain containing 1-like |
| chr13_+_23065500 | 2.97 |
ENSDART00000158370
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr6_-_12224562 | 2.95 |
ENSDART00000090266
ENSDART00000144028 |
gpd2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr18_-_36291570 | 2.92 |
|
|
|
| chr14_+_23220869 | 2.88 |
ENSDART00000112930
|
si:ch211-221f10.2
|
si:ch211-221f10.2 |
| chr9_-_22371512 | 2.79 |
ENSDART00000101809
|
crygm2d6
|
crystallin, gamma M2d6 |
| chr19_+_5375413 | 2.75 |
ENSDART00000141237
|
si:dkeyp-113d7.10
|
si:dkeyp-113d7.10 |
| chr15_+_5935952 | 2.71 |
ENSDART00000152520
ENSDART00000145827 |
sh3bgr
|
SH3 domain binding glutamate-rich protein |
| chr24_-_3334678 | 2.71 |
|
|
|
| chr11_-_38940966 | 2.71 |
ENSDART00000105133
|
wnt4a
|
wingless-type MMTV integration site family, member 4a |
| chr7_-_29300402 | 2.70 |
ENSDART00000099477
|
rorab
|
RAR-related orphan receptor A, paralog b |
| chr23_+_24999817 | 2.70 |
ENSDART00000137486
|
klhl21
|
kelch-like family member 21 |
| chr4_+_6024067 | 2.70 |
|
|
|
| chr3_+_28450576 | 2.69 |
ENSDART00000150893
|
sept12
|
septin 12 |
| chr13_-_2083873 | 2.67 |
ENSDART00000129773
|
mlip
|
muscular LMNA-interacting protein |
| chr7_+_38735582 | 2.65 |
ENSDART00000173735
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr5_+_51201924 | 2.65 |
ENSDART00000087467
|
cmya5
|
cardiomyopathy associated 5 |
| chr3_-_52419408 | 2.63 |
ENSDART00000154260
|
si:dkey-210j14.4
|
si:dkey-210j14.4 |
| chr24_-_6617860 | 2.60 |
ENSDART00000166216
|
arhgap21a
|
Rho GTPase activating protein 21a |
| chr16_-_13914368 | 2.56 |
|
|
|
| chr16_-_28943421 | 2.53 |
ENSDART00000149501
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr15_-_25581628 | 2.52 |
|
|
|
| chr19_-_7531649 | 2.51 |
ENSDART00000104750
|
mllt11
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 11 |
| chr12_+_23717554 | 2.48 |
ENSDART00000152997
|
svila
|
supervillin a |
| chr16_-_30816954 | 2.43 |
ENSDART00000129594
|
ptk2ab
|
protein tyrosine kinase 2ab |
| chr17_+_8874210 | 2.41 |
|
|
|
| chr3_+_40028301 | 2.41 |
ENSDART00000011568
|
syngr3a
|
synaptogyrin 3a |
| chr15_-_7327976 | 2.40 |
|
|
|
| chr3_+_32394550 | 2.39 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr5_-_57016269 | 2.39 |
ENSDART00000074264
|
crfb12
|
cytokine receptor family member B12 |
| chr25_-_18044103 | 2.38 |
ENSDART00000113581
|
kitlga
|
kit ligand a |
| chr3_-_5318289 | 2.37 |
ENSDART00000137105
|
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr3_+_28822408 | 2.36 |
ENSDART00000133528
|
lgals2a
|
lectin, galactoside-binding, soluble, 2a |
| chr18_+_40364675 | 2.36 |
ENSDART00000098791
ENSDART00000049171 |
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr23_+_17856104 | 2.35 |
ENSDART00000154427
|
CR381647.1
|
ENSDARG00000097211 |
| chr7_-_29300448 | 2.33 |
ENSDART00000019140
|
rorab
|
RAR-related orphan receptor A, paralog b |
| chr24_+_25326286 | 2.31 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| chr1_-_21987718 | 2.30 |
ENSDART00000128918
|
fgfbp1b
|
fibroblast growth factor binding protein 1b |
| chr16_+_24766043 | 2.29 |
ENSDART00000114304
|
ywhabl
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide like |
| chr3_+_31821397 | 2.29 |
ENSDART00000148861
|
kcnc3a
|
potassium voltage-gated channel, Shaw-related subfamily, member 3a |
| chr21_-_5692160 | 2.28 |
ENSDART00000075137
ENSDART00000151202 |
ccni
|
cyclin I |
| chr3_+_24587720 | 2.27 |
|
|
|
| chr5_-_63912916 | 2.25 |
|
|
|
| chr23_-_20026717 | 2.21 |
ENSDART00000153828
|
atp2b3b
|
ATPase, Ca++ transporting, plasma membrane 3b |
| chr2_-_3846844 | 2.19 |
ENSDART00000146861
|
mmp16b
|
matrix metallopeptidase 16b (membrane-inserted) |
| chr20_-_36672566 | 2.17 |
ENSDART00000062893
|
enah
|
enabled homolog (Drosophila) |
| chr11_+_18020191 | 2.14 |
ENSDART00000125984
|
mustn1b
|
musculoskeletal, embryonic nuclear protein 1b |
| chr23_-_18958008 | 2.14 |
ENSDART00000133419
|
CR847953.1
|
ENSDARG00000057403 |
| chr7_-_22685631 | 2.14 |
ENSDART00000122113
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr7_+_15629256 | 2.10 |
ENSDART00000138689
ENSDART00000137235 |
pax6b
|
paired box 6b |
| chr21_+_20347283 | 2.04 |
ENSDART00000026430
|
hspb11
|
heat shock protein, alpha-crystallin-related, b11 |
| chr3_+_22774002 | 2.04 |
|
|
|
| chr1_-_40557436 | 2.03 |
ENSDART00000144424
|
rgs12b
|
regulator of G protein signaling 12b |
| chr2_-_6351592 | 2.02 |
|
|
|
| chr5_+_36677034 | 2.02 |
ENSDART00000159402
|
si:dkey-17o15.2
|
si:dkey-17o15.2 |
| chr8_-_50270783 | 2.00 |
ENSDART00000146056
|
nkx3-1
|
NK3 homeobox 1 |
| chr24_+_25326233 | 1.99 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| chr6_-_52234513 | 1.94 |
ENSDART00000133554
|
tomm34
|
translocase of outer mitochondrial membrane 34 |
| chr1_+_5474009 | 1.92 |
ENSDART00000136060
|
prkag3a
|
protein kinase, AMP-activated, gamma 3a non-catalytic subunit |
| chr23_-_28286971 | 1.91 |
|
|
|
| chr9_-_28588288 | 1.89 |
ENSDART00000104317
ENSDART00000064343 |
klf7b
|
Kruppel-like factor 7b |
| chr12_+_5969088 | 1.88 |
ENSDART00000114961
|
si:ch211-131k2.3
|
si:ch211-131k2.3 |
| chr5_-_22847399 | 1.87 |
ENSDART00000169258
|
nlgn3b
|
neuroligin 3b |
| chr1_-_54532919 | 1.82 |
ENSDART00000111671
|
mri1
|
methylthioribose-1-phosphate isomerase 1 |
| KN150425v1_+_103569 | 1.81 |
ENSDART00000169530
|
cyp1c1
|
cytochrome P450, family 1, subfamily C, polypeptide 1 |
| chr10_-_37098396 | 1.81 |
ENSDART00000155277
|
BX323076.2
|
ENSDARG00000097288 |
| chr23_+_44137593 | 1.80 |
ENSDART00000148470
|
CU993818.1
|
ENSDARG00000095873 |
| chr10_-_17274395 | 1.79 |
ENSDART00000135679
|
rab36
|
RAB36, member RAS oncogene family |
| chr10_-_24401876 | 1.78 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr21_+_11592175 | 1.77 |
ENSDART00000147473
|
pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr14_-_32838287 | 1.76 |
|
|
|
| chr15_-_25581590 | 1.73 |
|
|
|
| chr17_-_5426195 | 1.72 |
ENSDART00000035944
|
clic5a
|
chloride intracellular channel 5a |
| chr25_-_23648033 | 1.70 |
ENSDART00000089278
|
si:ch211-236l14.4
|
si:ch211-236l14.4 |
| chr21_+_13031158 | 1.69 |
ENSDART00000081426
|
odf2a
|
outer dense fiber of sperm tails 2a |
| chr13_+_18130206 | 1.69 |
|
|
|
| chr20_-_2075861 | 1.68 |
ENSDART00000113237
|
ARHGAP18
|
Rho GTPase activating protein 18 |
| chr21_-_32449627 | 1.68 |
|
|
|
| chr20_-_26102193 | 1.65 |
ENSDART00000136518
|
capn3b
|
calpain 3b |
| chr20_-_26102273 | 1.64 |
ENSDART00000063177
|
capn3b
|
calpain 3b |
| chr5_-_56965438 | 1.63 |
ENSDART00000074306
|
hspb2
|
heat shock protein, alpha-crystallin-related, b2 |
| chr20_-_37673852 | 1.62 |
|
|
|
| chr3_-_42275696 | 1.61 |
ENSDART00000167844
|
mafk
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K |
| chr20_+_6640497 | 1.61 |
ENSDART00000138361
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr20_-_48255596 | 1.59 |
ENSDART00000031167
|
tfap2d
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
| chr9_-_43072092 | 1.58 |
ENSDART00000125953
|
ttn.1
|
titin, tandem duplicate 1 |
| chr25_+_33924376 | 1.53 |
|
|
|
| chr6_-_31268267 | 1.42 |
|
|
|
| chr2_-_16586136 | 1.40 |
ENSDART00000133708
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr24_-_32115473 | 1.38 |
ENSDART00000168419
|
rsu1
|
Ras suppressor protein 1 |
| chr3_+_32272097 | 1.38 |
ENSDART00000156982
|
si:ch211-195b15.8
|
si:ch211-195b15.8 |
| chr2_-_24113073 | 1.38 |
ENSDART00000148685
|
xirp1
|
xin actin binding repeat containing 1 |
| chr13_+_18130077 | 1.35 |
|
|
|
| chr22_-_37676556 | 1.35 |
ENSDART00000028085
|
ttc14
|
tetratricopeptide repeat domain 14 |
| chr6_+_42478185 | 1.33 |
ENSDART00000150226
|
mst1ra
|
macrophage stimulating 1 receptor a |
| chr14_+_14541725 | 1.33 |
ENSDART00000169932
|
ccdc142
|
coiled-coil domain containing 142 |
| chr5_-_63912962 | 1.32 |
|
|
|
| chr20_+_19879839 | 1.32 |
ENSDART00000132576
|
BX510657.1
|
ENSDARG00000093899 |
| chr16_+_34157866 | 1.30 |
ENSDART00000140552
|
tcea3
|
transcription elongation factor A (SII), 3 |
| chr6_+_27348529 | 1.30 |
|
|
|
| chr8_-_46903869 | 1.30 |
ENSDART00000161462
|
acot7
|
acyl-CoA thioesterase 7 |
| chr2_-_19532470 | 1.29 |
ENSDART00000166881
|
BX323588.1
|
ENSDARG00000102520 |
| chr2_-_10300329 | 1.27 |
ENSDART00000138081
|
bcl6ab
|
B-cell CLL/lymphoma 6a, genome duplicate b |
| chr3_+_29314649 | 1.25 |
ENSDART00000020381
|
grap2a
|
GRB2-related adaptor protein 2a |
| chr9_-_24220466 | 1.23 |
ENSDART00000021218
|
rpe
|
ribulose-5-phosphate-3-epimerase |
| chr19_+_15666408 | 1.22 |
ENSDART00000131134
|
si:ch211-206a7.2
|
si:ch211-206a7.2 |
| chr6_-_18814348 | 1.21 |
ENSDART00000115118
|
tns1a
|
tensin 1a |
| chr24_+_20815964 | 1.20 |
ENSDART00000133008
|
si:ch211-161h7.8
|
si:ch211-161h7.8 |
| chr20_-_16553010 | 1.18 |
|
|
|
| chr18_-_33072423 | 1.17 |
ENSDART00000177619
|
CU929403.2
|
ENSDARG00000106481 |
| chr1_-_54319671 | 1.16 |
ENSDART00000074058
|
slc25a23a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23a |
| chr7_-_18751216 | 1.16 |
ENSDART00000157618
|
HACD4
|
3-hydroxyacyl-CoA dehydratase 4 |
| chr7_+_57374714 | 1.15 |
ENSDART00000110623
|
arsj
|
arylsulfatase family, member J |
| chr7_-_29861232 | 1.14 |
|
|
|
| chr13_-_30807799 | 1.12 |
|
|
|
| chr3_-_49010673 | 1.12 |
ENSDART00000109544
|
si:ch1073-100f3.2
|
si:ch1073-100f3.2 |
| chr20_-_43281269 | 1.11 |
|
|
|
| chr23_-_27707182 | 1.10 |
ENSDART00000103639
|
arf3a
|
ADP-ribosylation factor 3a |
| chr2_-_49149839 | 1.10 |
|
|
|
| chr18_+_39345919 | 1.09 |
ENSDART00000012164
|
tmod2
|
tropomodulin 2 |
| chr24_+_41009372 | 1.08 |
ENSDART00000165952
|
CU633479.3
|
ENSDARG00000099523 |
| chr16_-_31517858 | 1.06 |
ENSDART00000145691
|
ENSDARG00000054814
|
ENSDARG00000054814 |
| chr6_-_52234640 | 1.05 |
ENSDART00000133554
|
tomm34
|
translocase of outer mitochondrial membrane 34 |
| chr11_+_25266623 | 1.04 |
ENSDART00000154213
|
tfe3b
|
transcription factor binding to IGHM enhancer 3b |
| chr25_+_20618512 | 1.04 |
ENSDART00000144748
|
ergic2
|
ERGIC and golgi 2 |
| chr18_+_40365208 | 1.02 |
ENSDART00000167134
|
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr25_+_3551924 | 1.02 |
ENSDART00000154348
|
prnprs3
|
prion protein, related sequence 3 |
| chr3_+_34063993 | 1.02 |
|
|
|
| chr3_+_22724101 | 1.01 |
ENSDART00000167127
|
BX322574.1
|
ENSDARG00000098353 |
| chr6_+_19476528 | 1.01 |
ENSDART00000113911
|
micall1a
|
MICAL-like 1a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 9.5 | GO:0051899 | regulation of mitochondrial membrane potential(GO:0051881) membrane depolarization(GO:0051899) regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 1.4 | 20.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 1.3 | 3.8 | GO:1990092 | calcium-dependent self proteolysis(GO:1990092) |
| 1.2 | 3.7 | GO:0097377 | interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 1.2 | 7.1 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 1.1 | 11.6 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 1.0 | 5.1 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.9 | 7.1 | GO:0046864 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.9 | 4.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.8 | 1.6 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.6 | 1.9 | GO:0097104 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.6 | 1.8 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.5 | 2.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.5 | 9.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.5 | 2.0 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.4 | 4.9 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.4 | 2.1 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.4 | 2.9 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.4 | 2.1 | GO:0090104 | pancreatic epsilon cell differentiation(GO:0090104) |
| 0.4 | 1.2 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.4 | 4.5 | GO:0007035 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.4 | 4.8 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.3 | 2.4 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.3 | 5.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.3 | 5.8 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.3 | 2.4 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.3 | 3.5 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.3 | 2.7 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.3 | 3.5 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.3 | 0.9 | GO:0099509 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.3 | 4.0 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.3 | 0.8 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.3 | 2.4 | GO:0070309 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.3 | 3.0 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.2 | 3.7 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.2 | 5.3 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 1.1 | GO:0015744 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.2 | 5.3 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.2 | 1.9 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.2 | 0.6 | GO:0045905 | positive regulation of translational termination(GO:0045905) |
| 0.2 | 0.6 | GO:0055004 | atrial cardiac myofibril assembly(GO:0055004) |
| 0.2 | 1.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 1.0 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.2 | 2.2 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.2 | 1.8 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 5.3 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.1 | 3.4 | GO:0021986 | habenula development(GO:0021986) |
| 0.1 | 1.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 1.3 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 0.5 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.1 | 2.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 1.0 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.1 | 1.3 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.1 | 3.2 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.1 | 1.9 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 2.4 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.1 | 3.0 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.1 | 2.4 | GO:0007599 | blood coagulation(GO:0007596) hemostasis(GO:0007599) |
| 0.1 | 3.4 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.9 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 1.8 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 4.3 | GO:0034765 | regulation of ion transmembrane transport(GO:0034765) |
| 0.0 | 6.5 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 3.2 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.0 | 0.9 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 2.2 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 3.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.5 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 3.3 | GO:0060047 | heart contraction(GO:0060047) |
| 0.0 | 3.1 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 1.3 | GO:0001817 | regulation of cytokine production(GO:0001817) |
| 0.0 | 1.0 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 1.0 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.5 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 2.9 | GO:0015711 | organic anion transport(GO:0015711) |
| 0.0 | 1.4 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 1.3 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.3 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.6 | GO:1901532 | regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 3.2 | GO:0006955 | immune response(GO:0006955) |
| 0.0 | 0.2 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.0 | 3.7 | GO:0006811 | ion transport(GO:0006811) |
| 0.0 | 2.7 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 0.5 | GO:0006414 | translational elongation(GO:0006414) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 8.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 1.0 | 11.1 | GO:0031430 | M band(GO:0031430) |
| 0.9 | 4.4 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.7 | 9.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.7 | 5.3 | GO:0031672 | A band(GO:0031672) |
| 0.6 | 4.5 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.6 | 3.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.5 | 5.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.5 | 2.3 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.4 | 3.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.4 | 4.9 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.3 | 12.1 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.2 | 5.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 6.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.2 | 1.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 2.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 2.7 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 3.0 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 2.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1.1 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.1 | 3.3 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.1 | 2.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 2.7 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 1.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 2.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 3.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.0 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.0 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 18.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 1.7 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 21.2 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 3.9 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.6 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 3.2 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 6.1 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 3.2 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 4.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.9 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.5 | GO:0005643 | nuclear pore(GO:0005643) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 8.6 | GO:0016936 | galactoside binding(GO:0016936) |
| 1.4 | 7.1 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 1.0 | 5.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 1.0 | 9.1 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.9 | 2.7 | GO:0005521 | lamin binding(GO:0005521) |
| 0.5 | 7.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.5 | 5.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.5 | 4.5 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.4 | 7.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.4 | 8.7 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.4 | 1.8 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.4 | 5.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.3 | 3.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.3 | 3.7 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.3 | 2.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 2.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 2.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 6.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 3.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.2 | 1.1 | GO:0015141 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) succinate transmembrane transporter activity(GO:0015141) |
| 0.2 | 3.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 1.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 4.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.2 | 5.0 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.1 | 6.8 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 2.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 3.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.5 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 0.1 | 0.5 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 72.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 1.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 1.8 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 1.2 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 1.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 2.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 4.8 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.1 | 3.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 1.9 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 3.0 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 1.8 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 1.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 3.0 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.1 | 5.0 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 2.4 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.9 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 2.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.7 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 2.9 | GO:0043492 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 3.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 2.6 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 2.3 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 12.6 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 1.1 | GO:0005126 | cytokine receptor binding(GO:0005126) |
| 0.0 | 1.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 2.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 4.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 2.7 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 1.3 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 1.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 2.6 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.5 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 5.5 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 1.5 | GO:0008134 | transcription factor binding(GO:0008134) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 12.6 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.5 | 4.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.2 | 4.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 3.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 2.8 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 5.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.2 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 2.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.6 | 9.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.5 | 3.2 | REACTOME ACETYLCHOLINE BINDING AND DOWNSTREAM EVENTS | Genes involved in Acetylcholine Binding And Downstream Events |
| 0.4 | 0.9 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.4 | 3.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.4 | 2.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.3 | 5.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 2.8 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.3 | 6.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 4.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 1.8 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 1.8 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.6 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 1.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 3.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |