Project
DANIO-CODE
Navigation
Downloads
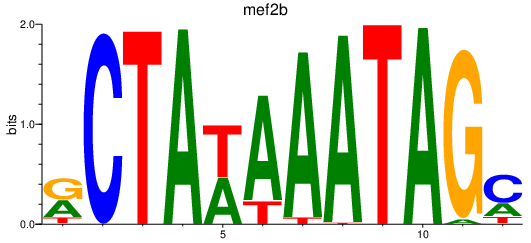
Results for mef2b
Z-value: 0.51
Transcription factors associated with mef2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mef2b
|
ENSDARG00000093170 | myocyte enhancer factor 2b |
Activity profile of mef2b motif
Sorted Z-values of mef2b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of mef2b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_60973097 | 2.19 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr3_-_32686790 | 2.05 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr5_-_71340996 | 1.33 |
ENSDART00000162526
|
smyd1a
|
SET and MYND domain containing 1a |
| chr9_-_33296340 | 1.30 |
ENSDART00000013918
|
casq2
|
calsequestrin 2 |
| chr15_-_27778111 | 1.29 |
ENSDART00000134373
|
lhx1a
|
LIM homeobox 1a |
| chr8_+_29953399 | 1.01 |
ENSDART00000149372
ENSDART00000007640 |
ptch1
ptch1
|
patched 1 patched 1 |
| chr11_+_11217547 | 1.00 |
ENSDART00000087105
|
myom2a
|
myomesin 2a |
| chr1_+_9999665 | 0.94 |
ENSDART00000054879
|
ENSDARG00000037679
|
ENSDARG00000037679 |
| chr6_-_42006033 | 0.91 |
ENSDART00000032527
|
cav3
|
caveolin 3 |
| chr24_-_31772736 | 0.90 |
|
|
|
| chr10_-_8074713 | 0.89 |
ENSDART00000140476
|
atp6v0a2a
|
ATPase, H+ transporting, lysosomal V0 subunit a2a |
| chr17_-_9869142 | 0.86 |
ENSDART00000008355
|
cfl2
|
cofilin 2 (muscle) |
| chr23_+_20183765 | 0.84 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr12_-_34614931 | 0.83 |
ENSDART00000152876
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr10_-_22834248 | 0.80 |
ENSDART00000079469
|
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr2_-_34010299 | 0.80 |
ENSDART00000140910
|
ptch2
|
patched 2 |
| chr23_-_39956151 | 0.73 |
ENSDART00000115330
|
ppp1r14c
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr20_+_15653121 | 0.70 |
ENSDART00000152702
|
jun
|
jun proto-oncogene |
| chr3_+_34064081 | 0.69 |
|
|
|
| chr10_-_22875997 | 0.66 |
ENSDART00000079454
|
vamp2
|
vesicle-associated membrane protein 2 |
| chr13_+_9100463 | 0.66 |
ENSDART00000058064
|
ENSDARG00000039726
|
ENSDARG00000039726 |
| chr1_-_5796394 | 0.64 |
ENSDART00000109356
|
klf7a
|
Kruppel-like factor 7a |
| chr2_+_20118917 | 0.63 |
ENSDART00000038648
|
ptbp2b
|
polypyrimidine tract binding protein 2b |
| chr19_-_31448000 | 0.62 |
ENSDART00000147504
|
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr1_-_16876855 | 0.61 |
ENSDART00000125125
|
acsl1a
|
acyl-CoA synthetase long-chain family member 1a |
| chr1_-_30772653 | 0.61 |
ENSDART00000087115
|
rims1b
|
regulating synaptic membrane exocytosis 1b |
| chr1_-_416138 | 0.60 |
ENSDART00000092524
|
rasa3
|
RAS p21 protein activator 3 |
| chr6_-_40724581 | 0.60 |
ENSDART00000035101
|
kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr10_-_24401876 | 0.58 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr14_+_23917724 | 0.58 |
ENSDART00000138082
ENSDART00000079215 |
stc2a
|
stanniocalcin 2a |
| chr25_-_22821132 | 0.56 |
|
|
|
| chr15_+_24064257 | 0.56 |
ENSDART00000156714
|
CR394545.1
|
ENSDARG00000097097 |
| chr23_-_7892862 | 0.55 |
ENSDART00000157612
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr15_-_14616083 | 0.54 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr21_+_19034242 | 0.54 |
ENSDART00000123309
|
nkx6.1
|
NK6 homeobox 1 |
| chr12_+_34018417 | 0.53 |
ENSDART00000032821
|
cyth1b
|
cytohesin 1b |
| chr10_-_37098396 | 0.53 |
ENSDART00000155277
|
BX323076.2
|
ENSDARG00000097288 |
| chr6_-_14010044 | 0.52 |
ENSDART00000149177
ENSDART00000174839 ENSDART00000089577 |
cacnb4b
|
calcium channel, voltage-dependent, beta 4b subunit |
| chr13_-_9737104 | 0.51 |
ENSDART00000158381
|
myom2b
|
myomesin 2b |
| chr21_+_11592175 | 0.50 |
ENSDART00000147473
|
pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr5_+_64411412 | 0.49 |
ENSDART00000158484
|
BX001023.2
|
ENSDARG00000101163 |
| chr24_+_25326286 | 0.49 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| chr17_-_14663139 | 0.48 |
ENSDART00000037371
|
ppp1r13ba
|
protein phosphatase 1, regulatory subunit 13Ba |
| chr6_-_42006225 | 0.48 |
ENSDART00000032527
|
cav3
|
caveolin 3 |
| chr24_+_25326233 | 0.48 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| chr20_+_41652149 | 0.47 |
ENSDART00000135715
|
fam184a
|
family with sequence similarity 184, member A |
| chr18_+_26737365 | 0.45 |
ENSDART00000141672
|
alpk3a
|
alpha-kinase 3a |
| chr5_-_51172369 | 0.44 |
ENSDART00000164267
ENSDART00000157866 |
homer1b
|
homer scaffolding protein 1b |
| chr7_-_23474701 | 0.44 |
ENSDART00000048050
|
ITGB1BP2
|
integrin subunit beta 1 binding protein 2 |
| chr22_+_26423320 | 0.43 |
ENSDART00000044085
|
ENSDARG00000034211
|
ENSDARG00000034211 |
| chr11_-_27455242 | 0.42 |
ENSDART00000045942
|
phf2
|
PHD finger protein 2 |
| chr9_-_28588288 | 0.42 |
ENSDART00000104317
ENSDART00000064343 |
klf7b
|
Kruppel-like factor 7b |
| chr11_-_27455348 | 0.41 |
ENSDART00000045942
|
phf2
|
PHD finger protein 2 |
| chr10_-_4979688 | 0.41 |
ENSDART00000093228
|
mat2al
|
methionine adenosyltransferase II, alpha-like |
| chr3_-_52419408 | 0.41 |
ENSDART00000154260
|
si:dkey-210j14.4
|
si:dkey-210j14.4 |
| chr21_-_12179911 | 0.40 |
ENSDART00000081510
ENSDART00000151297 |
celf4
|
CUGBP, Elav-like family member 4 |
| chr12_+_42244935 | 0.39 |
ENSDART00000166484
|
si:ch211-221j21.3
|
si:ch211-221j21.3 |
| chr1_+_11541423 | 0.39 |
ENSDART00000170760
|
tmod1
|
tropomodulin 1 |
| chr23_-_32231333 | 0.38 |
ENSDART00000000876
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr24_-_27800234 | 0.38 |
ENSDART00000105778
|
hs6st1b
|
heparan sulfate 6-O-sulfotransferase 1b |
| chr5_-_10445847 | 0.38 |
ENSDART00000163139
|
rtn4r
|
reticulon 4 receptor |
| chr11_-_38940966 | 0.37 |
ENSDART00000105133
|
wnt4a
|
wingless-type MMTV integration site family, member 4a |
| chr11_+_44922098 | 0.37 |
ENSDART00000172999
|
mafgb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
| chr20_-_26102273 | 0.35 |
ENSDART00000063177
|
capn3b
|
calpain 3b |
| chr5_+_64411584 | 0.35 |
ENSDART00000158484
|
BX001023.2
|
ENSDARG00000101163 |
| chr8_-_50270783 | 0.35 |
ENSDART00000146056
|
nkx3-1
|
NK3 homeobox 1 |
| chr23_-_38565787 | 0.34 |
|
|
|
| chr1_-_24486146 | 0.34 |
ENSDART00000144711
|
tmem154
|
transmembrane protein 154 |
| chr20_-_26142897 | 0.34 |
ENSDART00000140255
|
si:dkey-12h9.6
|
si:dkey-12h9.6 |
| chr12_+_30448812 | 0.33 |
ENSDART00000126064
|
si:ch211-28p3.4
|
si:ch211-28p3.4 |
| chr10_+_1954904 | 0.32 |
ENSDART00000177433
ENSDART00000178960 |
CABZ01070746.1
|
ENSDARG00000107761 |
| chr23_+_44137593 | 0.32 |
ENSDART00000148470
|
CU993818.1
|
ENSDARG00000095873 |
| chr21_+_10769107 | 0.32 |
|
|
|
| chr16_-_30698002 | 0.32 |
ENSDART00000146508
|
ldlrad4b
|
low density lipoprotein receptor class A domain containing 4b |
| chr16_+_17061069 | 0.31 |
ENSDART00000111074
|
si:ch211-120k19.1
|
si:ch211-120k19.1 |
| chr18_+_40364675 | 0.31 |
ENSDART00000098791
ENSDART00000049171 |
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr20_-_26102193 | 0.31 |
ENSDART00000136518
|
capn3b
|
calpain 3b |
| chr13_+_18130206 | 0.30 |
|
|
|
| chr17_-_12872503 | 0.29 |
ENSDART00000113713
|
fam177a1
|
family with sequence similarity 177, member A1 |
| chr24_-_32115473 | 0.29 |
ENSDART00000168419
|
rsu1
|
Ras suppressor protein 1 |
| chr12_-_26339002 | 0.29 |
ENSDART00000153214
|
synpo2lb
|
synaptopodin 2-like b |
| chr10_+_3461461 | 0.28 |
|
|
|
| chr12_+_5969088 | 0.28 |
ENSDART00000114961
|
si:ch211-131k2.3
|
si:ch211-131k2.3 |
| chr23_-_38565708 | 0.28 |
|
|
|
| chr7_+_38735582 | 0.27 |
ENSDART00000173735
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr1_-_43936717 | 0.26 |
ENSDART00000081835
|
slc43a1b
|
solute carrier family 43 (amino acid system L transporter), member 1b |
| chr3_-_8608620 | 0.26 |
|
|
|
| chr1_-_54319671 | 0.26 |
ENSDART00000074058
|
slc25a23a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23a |
| chr3_+_22774002 | 0.25 |
|
|
|
| chr20_-_31841557 | 0.23 |
|
|
|
| chr6_+_36965023 | 0.23 |
ENSDART00000028895
|
negr1
|
neuronal growth regulator 1 |
| chr15_-_12297979 | 0.22 |
ENSDART00000165159
|
dscaml1
|
Down syndrome cell adhesion molecule like 1 |
| chr20_+_34552759 | 0.22 |
ENSDART00000135789
|
mettl11b
|
methyltransferase like 11B |
| chr8_+_7882144 | 0.21 |
|
|
|
| chr9_+_24255064 | 0.21 |
ENSDART00000101577
ENSDART00000159324 ENSDART00000023196 ENSDART00000079689 ENSDART00000172743 ENSDART00000171577 |
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr6_-_42005949 | 0.21 |
ENSDART00000032527
|
cav3
|
caveolin 3 |
| chr4_+_13451169 | 0.20 |
ENSDART00000172552
|
dyrk2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr13_+_18130077 | 0.19 |
|
|
|
| chr1_-_43936611 | 0.19 |
ENSDART00000170912
|
slc43a1b
|
solute carrier family 43 (amino acid system L transporter), member 1b |
| chr5_+_63687529 | 0.19 |
ENSDART00000162246
|
plpp7
|
phospholipid phosphatase 7 (inactive) |
| chr25_+_33924376 | 0.19 |
|
|
|
| chr6_+_27348529 | 0.19 |
|
|
|
| chr12_+_28795950 | 0.18 |
ENSDART00000076572
|
rnf40
|
ring finger protein 40 |
| chr7_+_21593005 | 0.18 |
ENSDART00000159626
|
si:dkey-85k7.7
|
si:dkey-85k7.7 |
| chr24_-_4418454 | 0.18 |
ENSDART00000066835
|
fzd8a
|
frizzled class receptor 8a |
| chr1_+_44379602 | 0.18 |
ENSDART00000129819
|
si:ch211-151p13.8
|
si:ch211-151p13.8 |
| chr3_+_22724101 | 0.17 |
ENSDART00000167127
|
BX322574.1
|
ENSDARG00000098353 |
| chr5_+_3375612 | 0.17 |
|
|
|
| chr7_-_7186964 | 0.16 |
ENSDART00000102620
|
six7
|
SIX homeobox 7 |
| chr24_+_8596066 | 0.16 |
ENSDART00000113493
|
tmem14ca
|
transmembrane protein 14Ca |
| chr13_-_31246639 | 0.16 |
ENSDART00000026692
|
ubtd1a
|
ubiquitin domain containing 1a |
| chr21_-_32449627 | 0.16 |
|
|
|
| chr14_+_35560934 | 0.15 |
ENSDART00000105604
|
zgc:77938
|
zgc:77938 |
| chr20_-_37673852 | 0.15 |
|
|
|
| chr12_+_6411166 | 0.15 |
ENSDART00000152616
|
prkg1b
|
protein kinase, cGMP-dependent, type Ib |
| chr23_-_38565866 | 0.14 |
|
|
|
| chr11_-_26515468 | 0.14 |
|
|
|
| chr2_+_37992682 | 0.14 |
ENSDART00000179233
|
CR376839.1
|
ENSDARG00000108815 |
| chr12_+_26930895 | 0.14 |
ENSDART00000152975
|
msl1b
|
male-specific lethal 1 homolog b (Drosophila) |
| chr4_+_14659537 | 0.13 |
|
|
|
| chr14_+_14541725 | 0.12 |
ENSDART00000169932
|
ccdc142
|
coiled-coil domain containing 142 |
| chr9_+_30174202 | 0.12 |
ENSDART00000020743
|
cmss1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr15_+_20607816 | 0.11 |
ENSDART00000169941
|
sgsm2
|
small G protein signaling modulator 2 |
| chr25_+_3551924 | 0.10 |
ENSDART00000154348
|
prnprs3
|
prion protein, related sequence 3 |
| chr7_-_6222134 | 0.10 |
ENSDART00000173197
|
hist1h4l
|
histone 1, H4, like |
| chr20_-_26102318 | 0.10 |
ENSDART00000063177
|
capn3b
|
calpain 3b |
| chr6_-_6091628 | 0.09 |
ENSDART00000132350
|
rtn4a
|
reticulon 4a |
| chr1_-_33700550 | 0.09 |
|
|
|
| chr15_+_14925920 | 0.09 |
|
|
|
| chr7_-_29861232 | 0.09 |
|
|
|
| chr3_+_34063993 | 0.09 |
|
|
|
| chr8_+_30446599 | 0.08 |
ENSDART00000085894
|
pgm5
|
phosphoglucomutase 5 |
| chr9_+_50907029 | 0.08 |
|
|
|
| chr25_-_27279272 | 0.07 |
ENSDART00000148121
|
zgc:153935
|
zgc:153935 |
| chr13_-_36456350 | 0.07 |
ENSDART00000143682
|
lgals3a
|
lectin, galactoside binding soluble 3a |
| chr4_+_75484112 | 0.07 |
ENSDART00000174393
|
si:dkey-240n22.8
|
si:dkey-240n22.8 |
| chr11_-_22143133 | 0.06 |
ENSDART00000159681
|
tfeb
|
transcription factor EB |
| chr12_+_26931140 | 0.06 |
ENSDART00000152975
|
msl1b
|
male-specific lethal 1 homolog b (Drosophila) |
| chr5_+_36348848 | 0.05 |
|
|
|
| chr9_+_29860757 | 0.04 |
ENSDART00000015377
|
rnaseh2b
|
ribonuclease H2, subunit B |
| chr4_+_11465367 | 0.04 |
ENSDART00000008584
|
gdi2
|
GDP dissociation inhibitor 2 |
| chr10_+_4979788 | 0.02 |
|
|
|
| chr12_-_34614903 | 0.01 |
ENSDART00000152876
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr7_+_33186069 | 0.01 |
ENSDART00000164921
|
paqr5b
|
progestin and adipoQ receptor family member Vb |
| chr8_-_23250112 | 0.01 |
|
|
|
| chr16_-_20084901 | 0.00 |
ENSDART00000103586
ENSDART00000079159 |
hdac9b
|
histone deacetylase 9b |
| chr3_+_30962542 | 0.00 |
ENSDART00000153074
|
si:dkey-66i24.9
|
si:dkey-66i24.9 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0097377 | interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 0.4 | 1.8 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.3 | 1.3 | GO:0014808 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0014808) sarcoplasmic reticulum calcium ion transport(GO:0070296) calcium ion transport from endoplasmic reticulum to cytosol(GO:1903514) |
| 0.3 | 0.8 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.3 | 0.8 | GO:1990092 | calcium-dependent self proteolysis(GO:1990092) |
| 0.2 | 0.9 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.2 | 1.6 | GO:0001765 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 0.5 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 0.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 1.3 | GO:0030241 | skeletal muscle thin filament assembly(GO:0030240) skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 0.4 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.9 | GO:0007035 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.3 | GO:0060394 | SMAD protein complex assembly(GO:0007183) regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.2 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.1 | 0.7 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.5 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.4 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.6 | GO:0043363 | nucleate erythrocyte differentiation(GO:0043363) |
| 0.0 | 0.5 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.5 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.5 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.5 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.3 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.2 | GO:0046294 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.0 | 0.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.5 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.5 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.6 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.2 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.6 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.3 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.2 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.5 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.2 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 0.1 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.3 | GO:0031430 | M band(GO:0031430) |
| 0.3 | 1.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.6 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.1 | 0.9 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 1.6 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.4 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.7 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.8 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.8 | GO:0005119 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.2 | 1.6 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.6 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.4 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 0.9 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.4 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 0.4 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 0.8 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.6 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.7 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) cGMP binding(GO:0030553) |
| 0.0 | 0.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.3 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.5 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 1.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 0.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 1.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |