Project
DANIO-CODE
Navigation
Downloads
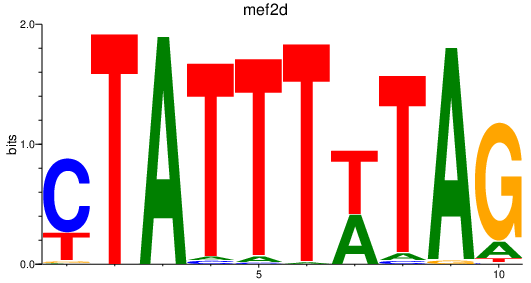
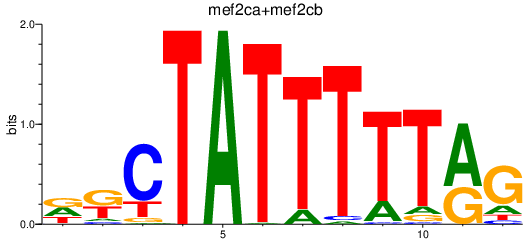
Results for mef2d_mef2ca+mef2cb
Z-value: 3.04
Transcription factors associated with mef2d_mef2ca+mef2cb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mef2d
|
ENSDARG00000040237 | myocyte enhancer factor 2d |
|
mef2cb
|
ENSDARG00000009418 | myocyte enhancer factor 2cb |
|
mef2ca
|
ENSDARG00000029764 | myocyte enhancer factor 2ca |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mef2ca | dr10_dc_chr10_-_43826919_43827049 | 0.96 | 1.9e-09 | Click! |
| mef2cb | dr10_dc_chr5_-_47634182_47634233 | 0.80 | 2.2e-04 | Click! |
| mef2d | dr10_dc_chr16_-_29229687_29229731 | 0.67 | 4.8e-03 | Click! |
Activity profile of mef2d_mef2ca+mef2cb motif
Sorted Z-values of mef2d_mef2ca+mef2cb motif
Network of associatons between targets according to the STRING database.
First level regulatory network of mef2d_mef2ca+mef2cb
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_6862901 | 16.06 |
ENSDART00000015732
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr3_-_60973097 | 16.03 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr12_-_4033069 | 15.84 |
ENSDART00000042200
|
aldoab
|
aldolase a, fructose-bisphosphate, b |
| chr9_-_33296340 | 11.68 |
ENSDART00000013918
|
casq2
|
calsequestrin 2 |
| chr5_-_71340996 | 9.97 |
ENSDART00000162526
|
smyd1a
|
SET and MYND domain containing 1a |
| chr6_+_52791515 | 9.81 |
ENSDART00000065682
|
matn4
|
matrilin 4 |
| chr15_+_24064257 | 8.30 |
ENSDART00000156714
|
CR394545.1
|
ENSDARG00000097097 |
| chr7_-_57941497 | 8.13 |
ENSDART00000114008
|
unm_hu7910
|
un-named hu7910 |
| chr6_+_3519697 | 8.04 |
ENSDART00000013588
|
klhl41b
|
kelch-like family member 41b |
| chr12_-_17590507 | 7.91 |
ENSDART00000010144
|
pvalb2
|
parvalbumin 2 |
| chr4_+_10065500 | 7.90 |
ENSDART00000026492
|
flncb
|
filamin C, gamma b (actin binding protein 280) |
| chr9_+_41720118 | 7.34 |
|
|
|
| chr23_+_44137593 | 7.12 |
ENSDART00000148470
|
CU993818.1
|
ENSDARG00000095873 |
| chr6_+_29420644 | 6.99 |
ENSDART00000065293
|
usp13
|
ubiquitin specific peptidase 13 (isopeptidase T-3) |
| chr10_-_22834248 | 6.85 |
ENSDART00000079469
|
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr24_+_20430778 | 6.62 |
ENSDART00000010488
|
klhl40b
|
kelch-like family member 40b |
| chr10_-_8074713 | 6.57 |
ENSDART00000140476
|
atp6v0a2a
|
ATPase, H+ transporting, lysosomal V0 subunit a2a |
| chr23_-_7283514 | 6.55 |
ENSDART00000156369
|
CR589876.1
|
ENSDARG00000096997 |
| chr5_+_66712197 | 6.52 |
ENSDART00000014822
|
ebf2
|
early B-cell factor 2 |
| chr24_+_36045759 | 6.05 |
ENSDART00000173322
|
obscnb
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF b |
| chr9_-_34491458 | 5.88 |
ENSDART00000049805
|
ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr2_+_27005381 | 5.75 |
|
|
|
| chr10_-_22875997 | 5.74 |
ENSDART00000079454
|
vamp2
|
vesicle-associated membrane protein 2 |
| chr20_-_9992898 | 5.73 |
ENSDART00000080664
|
ACTC1
|
zgc:86709 |
| chr3_+_34064081 | 5.62 |
|
|
|
| chr10_+_1954904 | 5.58 |
ENSDART00000177433
ENSDART00000178960 |
CABZ01070746.1
|
ENSDARG00000107761 |
| chr23_+_20183765 | 5.56 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr1_-_30772653 | 5.42 |
ENSDART00000087115
|
rims1b
|
regulating synaptic membrane exocytosis 1b |
| chr11_+_18020191 | 5.27 |
ENSDART00000125984
|
mustn1b
|
musculoskeletal, embryonic nuclear protein 1b |
| chr7_-_23474701 | 5.20 |
ENSDART00000048050
|
ITGB1BP2
|
integrin subunit beta 1 binding protein 2 |
| chr16_+_5466342 | 5.19 |
ENSDART00000160008
|
plecb
|
plectin b |
| chr25_-_30818836 | 5.03 |
ENSDART00000156828
|
prr33
|
proline rich 33 |
| chr21_+_19034242 | 5.02 |
ENSDART00000123309
|
nkx6.1
|
NK6 homeobox 1 |
| chr21_+_28408329 | 5.02 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr23_+_25005457 | 4.95 |
ENSDART00000136162
|
klhl21
|
kelch-like family member 21 |
| chr11_+_11217547 | 4.84 |
ENSDART00000087105
|
myom2a
|
myomesin 2a |
| chr4_-_9591451 | 4.73 |
ENSDART00000114060
|
cdnf
|
cerebral dopamine neurotrophic factor |
| chr20_+_15653121 | 4.72 |
ENSDART00000152702
|
jun
|
jun proto-oncogene |
| chr19_+_5156446 | 4.72 |
ENSDART00000151681
|
eno2
|
enolase 2 |
| chr9_-_7695437 | 4.67 |
ENSDART00000102715
|
tuba8l3
|
tubulin, alpha 8 like 3 |
| chr7_-_57727148 | 4.64 |
|
|
|
| chr12_-_17585587 | 4.63 |
ENSDART00000142427
|
pvalb3
|
parvalbumin 3 |
| chr16_+_34569479 | 4.59 |
ENSDART00000041007
|
stmn1b
|
stathmin 1b |
| chr11_-_27455348 | 4.56 |
ENSDART00000045942
|
phf2
|
PHD finger protein 2 |
| chr9_-_43340836 | 4.55 |
ENSDART00000059451
|
ccdc141
|
coiled-coil domain containing 141 |
| chr23_+_18796386 | 4.53 |
ENSDART00000137438
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr19_-_32900108 | 4.52 |
ENSDART00000050130
|
gmpr
|
guanosine monophosphate reductase |
| chr12_+_18403055 | 4.49 |
ENSDART00000090332
|
neurl2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr25_+_19992389 | 4.45 |
ENSDART00000143441
|
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr16_-_31963977 | 4.43 |
ENSDART00000027364
|
rbfox1l
|
RNA binding protein, fox-1 homolog (C. elegans) 1-like |
| chr19_-_41482388 | 4.43 |
ENSDART00000111982
|
sgce
|
sarcoglycan, epsilon |
| chr2_+_30932612 | 4.42 |
ENSDART00000132450
ENSDART00000137012 |
myom1a
|
myomesin 1a (skelemin) |
| chr10_+_32160464 | 4.38 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr14_-_20766448 | 4.36 |
ENSDART00000164146
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr24_+_25326286 | 4.33 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| chr18_+_26737365 | 4.29 |
ENSDART00000141672
|
alpk3a
|
alpha-kinase 3a |
| chr23_-_38565787 | 4.28 |
|
|
|
| chr24_-_23893617 | 4.26 |
ENSDART00000080549
|
lyz
|
lysozyme |
| chr3_+_28822408 | 4.25 |
ENSDART00000133528
|
lgals2a
|
lectin, galactoside-binding, soluble, 2a |
| chr19_+_41396014 | 4.23 |
ENSDART00000159444
ENSDART00000042990 ENSDART00000144544 |
col1a2
|
collagen, type I, alpha 2 |
| chr12_-_25973094 | 4.19 |
ENSDART00000171206
ENSDART00000171212 ENSDART00000170265 |
ldb3b
|
LIM domain binding 3b |
| chr9_-_37940101 | 4.19 |
ENSDART00000087663
|
sema5ba
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Ba |
| chr18_-_25190648 | 4.17 |
ENSDART00000175178
|
slco3a1
|
solute carrier organic anion transporter family, member 3A1 |
| chr21_+_11592175 | 4.16 |
ENSDART00000147473
|
pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr21_+_25899113 | 4.16 |
ENSDART00000141149
|
caln2
|
calneuron 2 |
| chr10_-_24401876 | 4.13 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr10_+_44364117 | 4.11 |
|
|
|
| chr23_-_38565708 | 4.08 |
|
|
|
| chr1_-_50066633 | 4.05 |
|
|
|
| chr21_-_25649117 | 3.97 |
ENSDART00000101205
|
phkg1b
|
phosphorylase kinase, gamma 1b (muscle) |
| chr20_-_39200252 | 3.95 |
ENSDART00000037318
ENSDART00000143379 |
rcan2
|
regulator of calcineurin 2 |
| chr20_-_10131480 | 3.90 |
ENSDART00000033976
|
meis2b
|
Meis homeobox 2b |
| chr7_+_31608828 | 3.90 |
ENSDART00000138491
|
mybpc3
|
myosin binding protein C, cardiac |
| chr17_+_38619041 | 3.87 |
ENSDART00000145147
|
sptb
|
spectrin, beta, erythrocytic |
| chr1_-_5796394 | 3.81 |
ENSDART00000109356
|
klf7a
|
Kruppel-like factor 7a |
| chr21_-_22694225 | 3.72 |
ENSDART00000101797
|
fbxo40.1
|
F-box protein 40, tandem duplicate 1 |
| chr18_+_20504980 | 3.69 |
ENSDART00000060295
|
rapsn
|
receptor-associated protein of the synapse, 43kD |
| chr9_+_6600592 | 3.68 |
ENSDART00000061577
|
fhl2a
|
four and a half LIM domains 2a |
| chr11_-_27455242 | 3.68 |
ENSDART00000045942
|
phf2
|
PHD finger protein 2 |
| chr15_-_9055873 | 3.67 |
ENSDART00000124998
|
rtn2a
|
reticulon 2a |
| chr24_+_25326233 | 3.59 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| chr1_-_43936717 | 3.59 |
ENSDART00000081835
|
slc43a1b
|
solute carrier family 43 (amino acid system L transporter), member 1b |
| chr11_-_18306222 | 3.54 |
ENSDART00000155474
|
fgd5a
|
FYVE, RhoGEF and PH domain containing 5a |
| chr14_-_29542922 | 3.48 |
|
|
|
| chr1_+_18272833 | 3.47 |
ENSDART00000132379
|
limch1a
|
LIM and calponin homology domains 1a |
| chr13_+_22119569 | 3.43 |
ENSDART00000108472
|
synpo2la
|
synaptopodin 2-like a |
| chr15_+_40161311 | 3.43 |
ENSDART00000063783
|
itm2ca
|
integral membrane protein 2Ca |
| chr10_+_1812271 | 3.37 |
|
|
|
| chr14_-_30025818 | 3.35 |
ENSDART00000007022
|
pdgfrl
|
platelet-derived growth factor receptor-like |
| chr10_-_37098396 | 3.35 |
ENSDART00000155277
|
BX323076.2
|
ENSDARG00000097288 |
| chr6_-_42006033 | 3.33 |
ENSDART00000032527
|
cav3
|
caveolin 3 |
| chr12_-_26339002 | 3.33 |
ENSDART00000153214
|
synpo2lb
|
synaptopodin 2-like b |
| chr11_-_38940966 | 3.32 |
ENSDART00000105133
|
wnt4a
|
wingless-type MMTV integration site family, member 4a |
| chr5_-_28090077 | 3.32 |
|
|
|
| chr18_+_40364675 | 3.31 |
ENSDART00000098791
ENSDART00000049171 |
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr5_-_22847399 | 3.27 |
ENSDART00000169258
|
nlgn3b
|
neuroligin 3b |
| chr18_+_26102262 | 3.26 |
ENSDART00000164495
|
znf710a
|
zinc finger protein 710a |
| chr15_+_19717045 | 3.23 |
ENSDART00000138680
|
lim2.3
|
lens intrinsic membrane protein 2.3 |
| chr13_+_7575753 | 3.19 |
|
|
|
| chr25_-_22821132 | 3.17 |
|
|
|
| chr9_-_28588288 | 3.11 |
ENSDART00000104317
ENSDART00000064343 |
klf7b
|
Kruppel-like factor 7b |
| chr12_+_22605268 | 3.09 |
|
|
|
| chr1_+_13386937 | 3.07 |
ENSDART00000021693
|
ank2a
|
ankyrin 2a, neuronal |
| chr3_-_39031305 | 3.06 |
ENSDART00000022393
|
si:dkeyp-57f11.2
|
si:dkeyp-57f11.2 |
| chr4_+_19545842 | 3.05 |
ENSDART00000140028
|
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr15_-_27778111 | 3.02 |
ENSDART00000134373
|
lhx1a
|
LIM homeobox 1a |
| chr3_+_18406137 | 2.96 |
ENSDART00000158791
|
cbx4
|
chromobox homolog 4 (Pc class homolog, Drosophila) |
| chr23_+_24999817 | 2.96 |
ENSDART00000137486
|
klhl21
|
kelch-like family member 21 |
| chr2_+_2895642 | 2.93 |
ENSDART00000032459
|
aqp1a.1
|
aquaporin 1a (Colton blood group), tandem duplicate 1 |
| chr24_+_16924647 | 2.89 |
ENSDART00000014787
|
pip4k2aa
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha a |
| chr21_+_10769107 | 2.88 |
|
|
|
| chr1_-_10391257 | 2.88 |
ENSDART00000102903
|
dmd
|
dystrophin |
| chr9_-_41982635 | 2.86 |
ENSDART00000144573
|
obsl1b
|
obscurin-like 1b |
| chr6_+_14822990 | 2.81 |
ENSDART00000149949
|
pou3f3b
|
POU class 3 homeobox 3b |
| chr7_+_31608878 | 2.80 |
ENSDART00000099789
|
mybpc3
|
myosin binding protein C, cardiac |
| chr7_-_13128257 | 2.77 |
ENSDART00000164326
|
si:ch73-119p20.1
|
si:ch73-119p20.1 |
| chr17_-_32913432 | 2.72 |
ENSDART00000077476
|
prox1a
|
prospero homeobox 1a |
| chr3_-_49010673 | 2.72 |
ENSDART00000109544
|
si:ch1073-100f3.2
|
si:ch1073-100f3.2 |
| chr13_-_30807799 | 2.71 |
|
|
|
| chr20_-_37673852 | 2.70 |
|
|
|
| chr23_-_5751064 | 2.70 |
ENSDART00000067351
|
tnnt2a
|
troponin T type 2a (cardiac) |
| chr24_+_26922969 | 2.66 |
|
|
|
| chr21_-_32449627 | 2.66 |
|
|
|
| chr15_-_14616083 | 2.64 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr19_-_24859287 | 2.63 |
ENSDART00000163763
|
thbs3b
|
thrombospondin 3b |
| chr3_+_18249107 | 2.62 |
ENSDART00000141100
ENSDART00000138107 |
rps2
|
ribosomal protein S2 |
| chr4_+_21996784 | 2.60 |
ENSDART00000040266
|
myf6
|
myogenic factor 6 |
| chr4_-_2615160 | 2.58 |
ENSDART00000140760
|
e2f7
|
E2F transcription factor 7 |
| chr25_+_16849382 | 2.58 |
ENSDART00000016591
|
fgf6a
|
fibroblast growth factor 6a |
| chr15_+_7179382 | 2.57 |
ENSDART00000101578
|
her8.2
|
hairy-related 8.2 |
| chr25_+_33924376 | 2.56 |
|
|
|
| chr1_-_43936611 | 2.55 |
ENSDART00000170912
|
slc43a1b
|
solute carrier family 43 (amino acid system L transporter), member 1b |
| chr6_+_14823185 | 2.55 |
ENSDART00000149202
|
pou3f3b
|
POU class 3 homeobox 3b |
| chr16_-_30698002 | 2.55 |
ENSDART00000146508
|
ldlrad4b
|
low density lipoprotein receptor class A domain containing 4b |
| chr3_+_59560881 | 2.54 |
ENSDART00000084738
|
ppp1r27a
|
protein phosphatase 1, regulatory subunit 27a |
| chr15_+_125263 | 2.54 |
|
|
|
| chr12_+_3043444 | 2.50 |
ENSDART00000149427
|
sgca
|
sarcoglycan, alpha |
| chr8_-_11191783 | 2.48 |
ENSDART00000131171
|
unc45b
|
unc-45 myosin chaperone B |
| chr16_+_34157866 | 2.46 |
ENSDART00000140552
|
tcea3
|
transcription elongation factor A (SII), 3 |
| chr8_+_10267246 | 2.45 |
ENSDART00000159312
ENSDART00000160766 |
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr7_-_29300448 | 2.42 |
ENSDART00000019140
|
rorab
|
RAR-related orphan receptor A, paralog b |
| chr13_+_22350043 | 2.42 |
ENSDART00000136863
|
ldb3a
|
LIM domain binding 3a |
| chr14_-_12001505 | 2.41 |
ENSDART00000171136
|
myot
|
myotilin |
| chr16_-_7963894 | 2.38 |
ENSDART00000172836
|
CABZ01061349.1
|
ENSDARG00000105487 |
| chr22_-_31110564 | 2.36 |
ENSDART00000022445
|
cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr7_+_38735582 | 2.32 |
ENSDART00000173735
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr14_-_23784679 | 2.32 |
ENSDART00000158576
|
msx2a
|
muscle segment homeobox 2a |
| chr12_+_25684420 | 2.30 |
ENSDART00000024415
|
epas1a
|
endothelial PAS domain protein 1a |
| chr12_-_48758684 | 2.29 |
ENSDART00000130190
|
uros
|
uroporphyrinogen III synthase |
| chr1_-_54319671 | 2.29 |
ENSDART00000074058
|
slc25a23a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23a |
| chr22_+_26423320 | 2.28 |
ENSDART00000044085
|
ENSDARG00000034211
|
ENSDARG00000034211 |
| chr7_-_39170016 | 2.27 |
ENSDART00000161191
|
BX842701.1
|
ENSDARG00000101872 |
| chr12_-_2905505 | 2.27 |
ENSDART00000157726
|
fam20a
|
family with sequence similarity 20, member A |
| chr13_+_7974393 | 2.26 |
ENSDART00000163103
|
galm
|
galactose mutarotase |
| chr1_-_21987718 | 2.26 |
ENSDART00000128918
|
fgfbp1b
|
fibroblast growth factor binding protein 1b |
| chr16_-_31517906 | 2.25 |
ENSDART00000145691
|
ENSDARG00000054814
|
ENSDARG00000054814 |
| chr5_+_38236956 | 2.24 |
ENSDART00000160236
|
fras1
|
Fraser extracellular matrix complex subunit 1 |
| KN150425v1_+_103569 | 2.23 |
ENSDART00000169530
|
cyp1c1
|
cytochrome P450, family 1, subfamily C, polypeptide 1 |
| chr3_-_29992171 | 2.23 |
ENSDART00000153562
|
ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr1_-_38097100 | 2.23 |
ENSDART00000148572
|
asb5b
|
ankyrin repeat and SOCS box containing 5b |
| chr3_+_22774002 | 2.21 |
|
|
|
| chr15_-_34987450 | 2.21 |
ENSDART00000009892
|
gabbr1a
|
gamma-aminobutyric acid (GABA) B receptor, 1a |
| chr2_-_24113073 | 2.16 |
ENSDART00000148685
|
xirp1
|
xin actin binding repeat containing 1 |
| chr23_-_39956151 | 2.16 |
ENSDART00000115330
|
ppp1r14c
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr5_-_66982187 | 2.15 |
|
|
|
| chr23_+_23559246 | 2.15 |
ENSDART00000172214
|
agrn
|
agrin |
| chr1_-_7417472 | 2.15 |
ENSDART00000161938
ENSDART00000163307 |
si:dkeyp-9d4.3
|
si:dkeyp-9d4.3 |
| chr13_-_31246639 | 2.14 |
ENSDART00000026692
|
ubtd1a
|
ubiquitin domain containing 1a |
| chr3_-_52419408 | 2.14 |
ENSDART00000154260
|
si:dkey-210j14.4
|
si:dkey-210j14.4 |
| chr3_-_29768364 | 2.13 |
ENSDART00000020311
|
rpl27
|
ribosomal protein L27 |
| chr21_-_12179911 | 2.12 |
ENSDART00000081510
ENSDART00000151297 |
celf4
|
CUGBP, Elav-like family member 4 |
| chr3_+_59491893 | 2.11 |
ENSDART00000084729
|
pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chr20_+_13279404 | 2.10 |
ENSDART00000025644
|
ppp2r5a
|
protein phosphatase 2, regulatory subunit B', alpha isoform |
| chr14_-_6822151 | 2.09 |
ENSDART00000171311
|
si:ch73-43g23.1
|
si:ch73-43g23.1 |
| chr24_-_31772736 | 2.08 |
|
|
|
| chr5_-_62592627 | 2.07 |
ENSDART00000131274
|
tbcelb
|
tubulin folding cofactor E-like b |
| chr21_+_10775209 | 2.07 |
|
|
|
| chr25_-_26292712 | 2.04 |
ENSDART00000067114
|
fbxl22
|
F-box and leucine-rich repeat protein 22 |
| chr22_+_16282153 | 2.03 |
ENSDART00000162685
ENSDART00000105678 |
lrrc39
|
leucine rich repeat containing 39 |
| chr8_+_17148864 | 2.02 |
ENSDART00000140531
|
dimt1l
|
DIM1 dimethyladenosine transferase 1-like (S. cerevisiae) |
| chr25_-_13057808 | 2.02 |
ENSDART00000172571
|
smpd3
|
sphingomyelin phosphodiesterase 3, neutral |
| chr9_-_30073641 | 2.02 |
|
|
|
| chr8_-_27667972 | 2.00 |
ENSDART00000086946
|
mov10b.1
|
Moloney leukemia virus 10b, tandem duplicate 1 |
| chr12_+_5969088 | 1.99 |
ENSDART00000114961
|
si:ch211-131k2.3
|
si:ch211-131k2.3 |
| chr22_-_6977038 | 1.99 |
ENSDART00000133143
ENSDART00000146813 |
gpd1b
|
glycerol-3-phosphate dehydrogenase 1b |
| chr12_+_22605229 | 1.99 |
|
|
|
| chr17_-_9869142 | 1.98 |
ENSDART00000008355
|
cfl2
|
cofilin 2 (muscle) |
| chr1_+_9999665 | 1.97 |
ENSDART00000054879
|
ENSDARG00000037679
|
ENSDARG00000037679 |
| chr4_+_14659537 | 1.96 |
|
|
|
| chr20_+_41652149 | 1.95 |
ENSDART00000135715
|
fam184a
|
family with sequence similarity 184, member A |
| chr24_-_21750082 | 1.94 |
|
|
|
| chr2_-_43318809 | 1.93 |
ENSDART00000132588
|
crema
|
cAMP responsive element modulator a |
| chr6_+_27348529 | 1.93 |
|
|
|
| chr11_+_44922098 | 1.93 |
ENSDART00000172999
|
mafgb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
| chr20_+_40248154 | 1.93 |
ENSDART00000121818
|
trdn
|
triadin |
| chr24_+_9315953 | 1.92 |
|
|
|
| chr1_+_51706536 | 1.91 |
|
|
|
| chr9_+_17431036 | 1.90 |
ENSDART00000140852
|
rgcc
|
regulator of cell cycle |
| chr1_+_11615117 | 1.89 |
ENSDART00000165733
|
gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr1_-_41281418 | 1.88 |
ENSDART00000014678
|
adra1d
|
adrenoceptor alpha 1D |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 11.7 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0014808) sarcoplasmic reticulum calcium ion transport(GO:0070296) calcium ion transport from endoplasmic reticulum to cytosol(GO:1903514) |
| 2.7 | 8.2 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 2.7 | 8.1 | GO:0042264 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 2.2 | 6.6 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 1.7 | 6.7 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 1.3 | 2.7 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 1.3 | 3.9 | GO:1990092 | calcium-dependent self proteolysis(GO:1990092) |
| 1.2 | 3.6 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 1.2 | 4.7 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 1.1 | 4.4 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 1.1 | 3.3 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 1.1 | 5.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 1.1 | 5.3 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 1.0 | 3.0 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 1.0 | 3.0 | GO:0097376 | interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 1.0 | 10.0 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) skeletal muscle myosin thick filament assembly(GO:0030241) |
| 1.0 | 13.9 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 1.0 | 2.9 | GO:0003091 | renal water homeostasis(GO:0003091) renal water transport(GO:0003097) carbon dioxide transport(GO:0015670) |
| 1.0 | 5.7 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.9 | 2.6 | GO:0032877 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.8 | 15.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.8 | 2.4 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.8 | 2.3 | GO:0097186 | enamel mineralization(GO:0070166) amelogenesis(GO:0097186) |
| 0.7 | 4.2 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.7 | 3.4 | GO:0010561 | negative regulation of glycoprotein biosynthetic process(GO:0010561) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.7 | 7.9 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.7 | 3.9 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.6 | 1.9 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.6 | 1.9 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.6 | 1.9 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.6 | 4.3 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.6 | 6.6 | GO:0007035 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.6 | 10.6 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.6 | 8.0 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.6 | 2.3 | GO:0019320 | hexose catabolic process(GO:0019320) galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.5 | 1.6 | GO:0010712 | regulation of collagen metabolic process(GO:0010712) regulation of granulocyte differentiation(GO:0030852) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) regulation of neutrophil differentiation(GO:0045658) |
| 0.5 | 2.7 | GO:0015743 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.5 | 10.6 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.5 | 2.5 | GO:0010991 | SMAD protein complex assembly(GO:0007183) regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.5 | 1.5 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) cellular response to vitamin D(GO:0071305) |
| 0.5 | 2.0 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.5 | 5.7 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.5 | 4.6 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.5 | 1.4 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) abducens nucleus development(GO:0021742) |
| 0.4 | 10.3 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.4 | 0.9 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.4 | 1.7 | GO:0050687 | negative regulation of response to biotic stimulus(GO:0002832) negative regulation of defense response to virus(GO:0050687) |
| 0.4 | 2.0 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.4 | 1.2 | GO:0042423 | catechol-containing compound biosynthetic process(GO:0009713) catecholamine biosynthetic process(GO:0042423) |
| 0.4 | 1.1 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.4 | 4.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.4 | 1.9 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.4 | 1.5 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.4 | 4.4 | GO:0000272 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.4 | 1.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.3 | 2.0 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.3 | 5.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.3 | 3.8 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.3 | 2.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.3 | 1.2 | GO:0031646 | positive regulation of myelination(GO:0031643) positive regulation of neurological system process(GO:0031646) |
| 0.3 | 6.6 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.3 | 2.9 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.3 | 10.7 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.3 | 5.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.3 | 1.4 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.3 | 4.7 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.3 | 2.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.3 | 2.0 | GO:0086003 | cardiac muscle cell contraction(GO:0086003) |
| 0.2 | 1.7 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 4.5 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.2 | 2.0 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.2 | 3.6 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.2 | 4.6 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.2 | 2.7 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.2 | 1.7 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.2 | 3.4 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.2 | 0.6 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.2 | 4.2 | GO:0009612 | response to mechanical stimulus(GO:0009612) |
| 0.2 | 1.3 | GO:0036065 | fucosylation(GO:0036065) |
| 0.2 | 5.9 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.2 | 1.6 | GO:0048512 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.2 | 1.1 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.2 | 4.0 | GO:0044773 | mitotic DNA damage checkpoint(GO:0044773) |
| 0.2 | 0.8 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 0.5 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.2 | 1.1 | GO:0048681 | regulation of myelination(GO:0031641) negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.1 | 2.2 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 2.9 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.1 | 8.7 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.1 | 2.1 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 1.0 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 1.9 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 1.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 8.6 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.1 | 2.0 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 1.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 2.2 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.1 | 0.1 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 4.0 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 4.1 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 1.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 3.2 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 2.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 1.2 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 1.0 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 0.5 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 0.3 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.1 | 1.8 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 | 0.8 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.1 | 0.6 | GO:0060088 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 1.9 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.1 | 3.3 | GO:0043242 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) negative regulation of protein complex disassembly(GO:0043242) actin filament capping(GO:0051693) regulation of protein depolymerization(GO:1901879) negative regulation of protein depolymerization(GO:1901880) |
| 0.1 | 4.6 | GO:0042552 | myelination(GO:0042552) |
| 0.1 | 0.4 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.1 | 0.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 2.3 | GO:0071456 | cellular response to decreased oxygen levels(GO:0036294) cellular response to oxygen levels(GO:0071453) cellular response to hypoxia(GO:0071456) |
| 0.1 | 1.1 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.5 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 1.5 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.1 | 3.6 | GO:0050808 | synapse organization(GO:0050808) |
| 0.1 | 1.8 | GO:0048634 | regulation of muscle organ development(GO:0048634) |
| 0.1 | 1.9 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.1 | 4.3 | GO:0048469 | cell maturation(GO:0048469) |
| 0.1 | 1.2 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.4 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 2.8 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.1 | 1.6 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 0.4 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.0 | 0.7 | GO:0015780 | nucleotide-sugar transport(GO:0015780) pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 1.8 | GO:0003146 | heart jogging(GO:0003146) |
| 0.0 | 1.3 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 2.7 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 3.8 | GO:0007492 | endoderm development(GO:0007492) |
| 0.0 | 1.3 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 1.6 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 2.6 | GO:0044344 | fibroblast growth factor receptor signaling pathway(GO:0008543) cellular response to fibroblast growth factor stimulus(GO:0044344) response to fibroblast growth factor(GO:0071774) |
| 0.0 | 2.4 | GO:0007626 | locomotory behavior(GO:0007626) |
| 0.0 | 1.7 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 1.1 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 1.3 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.6 | GO:0052646 | alditol phosphate metabolic process(GO:0052646) |
| 0.0 | 0.2 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.0 | 1.6 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.2 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.6 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 0.6 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 3.0 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.3 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.7 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 1.0 | GO:0001817 | regulation of cytokine production(GO:0001817) |
| 0.0 | 1.1 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 1.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:1901888 | regulation of cell junction assembly(GO:1901888) |
| 0.0 | 0.5 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 1.1 | GO:0033333 | fin development(GO:0033333) |
| 0.0 | 1.3 | GO:0006874 | cellular calcium ion homeostasis(GO:0006874) |
| 0.0 | 0.6 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 0.6 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.5 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.8 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.0 | 0.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 35.2 | GO:0031430 | M band(GO:0031430) |
| 2.3 | 11.7 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 1.5 | 4.5 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 1.4 | 4.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 1.2 | 4.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 1.2 | 6.9 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 1.1 | 9.1 | GO:0031672 | A band(GO:0031672) |
| 1.1 | 5.4 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.8 | 3.4 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.8 | 6.6 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.8 | 3.9 | GO:0008091 | spectrin(GO:0008091) |
| 0.7 | 3.0 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.7 | 2.1 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.6 | 13.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.5 | 10.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.5 | 5.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.4 | 1.3 | GO:0008352 | katanin complex(GO:0008352) |
| 0.4 | 2.2 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.4 | 4.7 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.3 | 1.4 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.3 | 2.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.3 | 7.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.3 | 1.6 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.3 | 17.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.3 | 1.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.2 | 2.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 5.7 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.2 | 1.9 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.2 | 8.2 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.2 | 6.0 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 3.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.5 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 2.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 2.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 1.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 4.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 2.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 2.0 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 1.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 2.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.8 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.1 | 1.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 1.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 2.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 11.6 | GO:0045202 | synapse(GO:0045202) |
| 0.1 | 19.9 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 2.2 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.7 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.0 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 1.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 2.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 4.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 17.6 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 3.0 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.8 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 15.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 1.5 | 4.5 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 1.4 | 4.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 1.3 | 10.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 1.2 | 3.6 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 1.2 | 4.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 1.1 | 4.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 1.0 | 5.0 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 1.0 | 2.9 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) glycerol transmembrane transporter activity(GO:0015168) |
| 0.9 | 6.0 | GO:0071253 | connexin binding(GO:0071253) |
| 0.8 | 4.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.8 | 4.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.7 | 6.6 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.6 | 2.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.6 | 8.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.5 | 2.7 | GO:0015140 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) succinate transmembrane transporter activity(GO:0015141) |
| 0.5 | 3.7 | GO:0043495 | protein anchor(GO:0043495) |
| 0.5 | 5.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.5 | 1.9 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) alpha1-adrenergic receptor activity(GO:0004937) |
| 0.5 | 1.9 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.5 | 6.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.4 | 1.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.4 | 2.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.4 | 1.7 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.4 | 1.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.4 | 2.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.4 | 4.5 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.4 | 2.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.4 | 3.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.3 | 2.7 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.3 | 1.7 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.3 | 1.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.3 | 6.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.3 | 1.6 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.3 | 5.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.3 | 4.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.3 | 0.8 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) cGMP binding(GO:0030553) |
| 0.3 | 5.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.3 | 2.6 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.2 | 1.4 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.2 | 3.0 | GO:0043236 | laminin binding(GO:0043236) |
| 0.2 | 2.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 1.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.2 | 8.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 3.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 1.8 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.2 | 5.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.2 | 6.6 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.2 | 10.3 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.2 | 1.0 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.2 | 8.1 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.2 | 1.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 3.4 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.2 | 1.1 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.2 | 2.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 2.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.2 | 4.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.2 | 2.0 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 2.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 87.9 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.2 | 2.9 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.2 | 0.5 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.2 | 3.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 1.8 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 1.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 1.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 2.2 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 2.6 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 1.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 3.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 5.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 2.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 1.5 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 6.9 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 0.9 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 27.5 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 3.3 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.1 | 6.1 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.1 | 0.5 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 1.3 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 3.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 2.7 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 1.5 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 3.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.8 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.7 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 2.3 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 1.3 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 1.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 5.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.5 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 2.0 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 2.2 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 1.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 5.3 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 1.2 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 8.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.7 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 1.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.6 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.1 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 1.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 15.5 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 7.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.4 | 6.0 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.2 | 15.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.2 | 14.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 1.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 2.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 4.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 2.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 1.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 2.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 0.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 1.5 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 2.0 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 1.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 4.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.7 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 1.4 | 4.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.6 | 4.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.6 | 5.0 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.6 | 1.9 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.6 | 3.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.5 | 1.0 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.5 | 3.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.4 | 4.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.3 | 4.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 4.8 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.2 | 3.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.2 | 1.6 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.2 | 4.4 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 1.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 1.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 3.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 2.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 2.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.4 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.2 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |