Project
DANIO-CODE
Navigation
Downloads
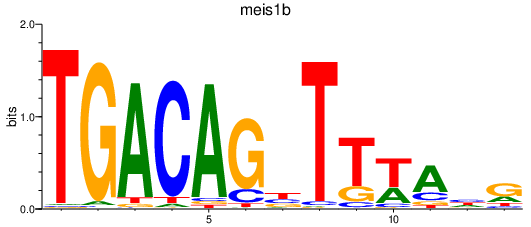
Results for meis1b
Z-value: 0.77
Transcription factors associated with meis1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
meis1b
|
ENSDARG00000012078 | Meis homeobox 1 b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| meis1b | dr10_dc_chr13_-_5440923_5440998 | -0.37 | 1.6e-01 | Click! |
Activity profile of meis1b motif
Sorted Z-values of meis1b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of meis1b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_34263554 | 1.05 |
ENSDART00000161290
|
pex1
|
peroxisomal biogenesis factor 1 |
| chr5_+_3567992 | 1.04 |
ENSDART00000129329
|
rpain
|
RPA interacting protein |
| chr20_+_35305119 | 0.90 |
ENSDART00000045135
|
fbxo16
|
F-box protein 16 |
| chr7_-_19116999 | 0.90 |
ENSDART00000165680
|
ntn4
|
netrin 4 |
| chr24_+_9887575 | 0.89 |
ENSDART00000172773
|
si:ch211-146l10.8
|
si:ch211-146l10.8 |
| chr15_+_31709362 | 0.86 |
ENSDART00000159634
|
b3glcta
|
beta 3-glucosyltransferase a |
| chr2_+_15432208 | 0.86 |
ENSDART00000027171
|
f3b
|
coagulation factor IIIb |
| chr23_-_27579257 | 0.78 |
ENSDART00000137229
|
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr25_+_19572283 | 0.76 |
ENSDART00000073472
|
zgc:113426
|
zgc:113426 |
| chr10_-_45182378 | 0.75 |
ENSDART00000161815
|
polm
|
polymerase (DNA directed), mu |
| chr2_+_15432130 | 0.75 |
ENSDART00000027171
|
f3b
|
coagulation factor IIIb |
| chr7_+_54991499 | 0.73 |
ENSDART00000073555
|
ctu2
|
cytosolic thiouridylase subunit 2 homolog (S. pombe) |
| chr24_+_32295105 | 0.73 |
ENSDART00000143570
|
CU207275.1
|
ENSDARG00000092965 |
| chr20_-_38884093 | 0.73 |
ENSDART00000153430
|
dnajc5ga
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a |
| chr4_+_16020464 | 0.72 |
ENSDART00000144611
|
CR749763.5
|
ENSDARG00000093983 |
| chr25_-_16455541 | 0.69 |
ENSDART00000125836
|
si:ch211-266k8.4
|
si:ch211-266k8.4 |
| chr9_+_4382041 | 0.68 |
|
|
|
| chr2_+_19588034 | 0.67 |
ENSDART00000163875
|
cc2d1b
|
coiled-coil and C2 domain containing 1B |
| chr5_-_66982706 | 0.66 |
ENSDART00000013605
|
zbtb20
|
zinc finger and BTB domain containing 20 |
| chr17_+_23709781 | 0.65 |
ENSDART00000034913
|
zgc:91976
|
zgc:91976 |
| chr13_-_32595706 | 0.64 |
ENSDART00000012232
|
pdss2
|
prenyl (decaprenyl) diphosphate synthase, subunit 2 |
| chr7_+_15781933 | 0.61 |
ENSDART00000161669
|
immp1l
|
inner mitochondrial membrane peptidase subunit 1 |
| chr24_-_12685892 | 0.61 |
ENSDART00000039312
|
ipo4
|
importin 4 |
| chr22_+_26840517 | 0.59 |
ENSDART00000158756
|
crebbpa
|
CREB binding protein a |
| chr12_+_46242558 | 0.59 |
ENSDART00000167510
|
hid1b
|
HID1 domain containing b |
| chr16_-_13723295 | 0.58 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr6_-_42973344 | 0.58 |
ENSDART00000150508
|
arl8ba
|
ADP-ribosylation factor-like 8Ba |
| chr24_-_19574944 | 0.57 |
ENSDART00000135405
|
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr23_+_23256896 | 0.56 |
ENSDART00000132296
|
klhl17
|
kelch-like family member 17 |
| chr6_-_8225364 | 0.56 |
|
|
|
| chr8_-_41511380 | 0.56 |
ENSDART00000019858
|
golga1
|
golgin A1 |
| chr16_-_13723352 | 0.55 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr11_-_2437396 | 0.55 |
|
|
|
| chr9_-_21287478 | 0.53 |
ENSDART00000018570
|
wars2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr12_-_4212149 | 0.53 |
ENSDART00000059298
|
zgc:92313
|
zgc:92313 |
| chr25_+_32114026 | 0.52 |
ENSDART00000156145
|
scaper
|
S-phase cyclin A-associated protein in the ER |
| chr5_+_40896320 | 0.52 |
ENSDART00000039369
|
fancg
|
Fanconi anemia, complementation group G |
| chr5_-_47609402 | 0.51 |
|
|
|
| chr17_+_23536720 | 0.51 |
ENSDART00000143508
|
pank1a
|
pantothenate kinase 1a |
| chr8_-_31375316 | 0.50 |
ENSDART00000159878
ENSDART00000164134 |
creb3l3l
|
cAMP responsive element binding protein 3-like 3 like |
| chr3_+_39437497 | 0.50 |
ENSDART00000055170
|
cln3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr24_+_32295150 | 0.50 |
ENSDART00000143570
|
CU207275.1
|
ENSDARG00000092965 |
| chr10_-_24796421 | 0.49 |
ENSDART00000064463
|
timm10b
|
translocase of inner mitochondrial membrane 10 homolog B (yeast) |
| chr6_-_24104127 | 0.49 |
ENSDART00000170915
|
si:ch73-389b16.2
|
si:ch73-389b16.2 |
| chr22_+_35299551 | 0.48 |
ENSDART00000165353
|
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr5_-_47609309 | 0.48 |
|
|
|
| chr24_+_26223767 | 0.48 |
|
|
|
| chr16_-_33864015 | 0.48 |
|
|
|
| chr2_-_32403565 | 0.47 |
ENSDART00000056634
|
ubtfl
|
upstream binding transcription factor, like |
| chr14_+_29601046 | 0.46 |
ENSDART00000143763
|
fam149a
|
family with sequence similarity 149 member A |
| chr17_-_21029294 | 0.46 |
ENSDART00000155300
|
bicc1a
|
BicC family RNA binding protein 1a |
| chr5_-_19459809 | 0.46 |
ENSDART00000148146
|
si:dkey-234h16.7
|
si:dkey-234h16.7 |
| chr5_-_19459698 | 0.46 |
ENSDART00000148146
|
si:dkey-234h16.7
|
si:dkey-234h16.7 |
| chr19_+_24483983 | 0.46 |
ENSDART00000141351
|
pex11b
|
peroxisomal biogenesis factor 11 beta |
| chr17_+_39794317 | 0.46 |
ENSDART00000154996
|
si:dkey-229e3.2
|
si:dkey-229e3.2 |
| chr11_-_26819599 | 0.44 |
ENSDART00000043091
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr10_+_29883772 | 0.44 |
ENSDART00000100032
|
hyou1
|
hypoxia up-regulated 1 |
| chr12_-_31346234 | 0.43 |
ENSDART00000175929
|
acsl5
|
acyl-CoA synthetase long-chain family member 5 |
| chr13_-_32595667 | 0.43 |
ENSDART00000012232
|
pdss2
|
prenyl (decaprenyl) diphosphate synthase, subunit 2 |
| chr5_+_40896478 | 0.43 |
ENSDART00000039369
|
fancg
|
Fanconi anemia, complementation group G |
| chr3_-_44466637 | 0.43 |
ENSDART00000169923
|
ndel1b
|
nudE neurodevelopment protein 1-like 1b |
| chr1_-_11191824 | 0.43 |
ENSDART00000163971
ENSDART00000123431 |
iqce
|
IQ motif containing E |
| chr17_+_24790812 | 0.42 |
ENSDART00000082251
|
spdya
|
speedy/RINGO cell cycle regulator family member A |
| chr8_-_31374705 | 0.41 |
ENSDART00000162872
|
creb3l3l
|
cAMP responsive element binding protein 3-like 3 like |
| chr4_-_7867294 | 0.41 |
ENSDART00000167943
|
nudt5
|
nudix (nucleoside diphosphate linked moiety X)-type motif 5 |
| chr3_+_56882443 | 0.41 |
|
|
|
| chr16_-_39291647 | 0.41 |
ENSDART00000171342
|
tmem42a
|
transmembrane protein 42a |
| chr20_+_37592206 | 0.40 |
ENSDART00000153092
|
BX088686.1
|
ENSDARG00000096774 |
| chr17_-_21029377 | 0.40 |
ENSDART00000155300
|
bicc1a
|
BicC family RNA binding protein 1a |
| chr8_-_1601007 | 0.39 |
ENSDART00000159705
|
dgcr6
|
DiGeorge syndrome critical region gene 6 |
| chr12_-_10384031 | 0.39 |
ENSDART00000052004
|
zgc:153595
|
zgc:153595 |
| chr18_+_45866556 | 0.39 |
ENSDART00000024615
|
rnpepl1
|
arginyl aminopeptidase like 1 |
| chr24_-_9874069 | 0.39 |
ENSDART00000176344
|
si:ch211-146l10.7
|
si:ch211-146l10.7 |
| chr13_-_33096999 | 0.39 |
ENSDART00000057386
|
golga5
|
golgin A5 |
| chr4_-_13025193 | 0.38 |
ENSDART00000177894
|
BX119963.3
|
ENSDARG00000106390 |
| chr12_-_10383960 | 0.38 |
ENSDART00000052004
|
zgc:153595
|
zgc:153595 |
| chr22_+_26840870 | 0.38 |
ENSDART00000166775
|
crebbpa
|
CREB binding protein a |
| chr17_+_34238753 | 0.37 |
ENSDART00000014306
|
mpp5a
|
membrane protein, palmitoylated 5a (MAGUK p55 subfamily member 5) |
| chr17_-_43542320 | 0.37 |
ENSDART00000154595
|
si:dkey-21a6.5
|
si:dkey-21a6.5 |
| chr3_+_56882467 | 0.37 |
|
|
|
| chr20_+_3321716 | 0.36 |
ENSDART00000108955
|
mfsd4b
|
major facilitator superfamily domain containing 4B |
| chr5_+_50602176 | 0.36 |
ENSDART00000159571
|
msh3
|
mutS homolog 3 (E. coli) |
| chr8_-_34509255 | 0.36 |
ENSDART00000112854
|
gapvd1
|
GTPase activating protein and VPS9 domains 1 |
| chr3_+_39437613 | 0.36 |
ENSDART00000055170
|
cln3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr9_-_25555518 | 0.36 |
ENSDART00000021672
|
epc2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr7_-_19274727 | 0.36 |
ENSDART00000114203
|
man2b2
|
mannosidase, alpha, class 2B, member 2 |
| chr12_-_31346300 | 0.35 |
ENSDART00000175929
|
acsl5
|
acyl-CoA synthetase long-chain family member 5 |
| chr15_+_14442857 | 0.35 |
ENSDART00000160145
|
si:ch211-11n16.2
|
si:ch211-11n16.2 |
| chr12_+_34790513 | 0.35 |
ENSDART00000015643
|
tbcc
|
tubulin folding cofactor C |
| chr3_+_37673971 | 0.35 |
|
|
|
| chr19_-_9584480 | 0.35 |
ENSDART00000091615
|
iffo1a
|
intermediate filament family orphan 1a |
| chr3_+_15605991 | 0.34 |
ENSDART00000140160
|
BX914211.1
|
ENSDARG00000094116 |
| chr17_-_24848180 | 0.34 |
ENSDART00000027957
|
hmgcl
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| chr9_-_25555313 | 0.34 |
ENSDART00000021672
|
epc2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr5_+_37144525 | 0.34 |
ENSDART00000148766
|
dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr21_+_22293328 | 0.34 |
ENSDART00000157839
|
nadk2
|
NAD kinase 2, mitochondrial |
| chr7_+_23735703 | 0.34 |
ENSDART00000033755
|
homezb
|
homeobox and leucine zipper encoding b |
| chr7_-_25252260 | 0.34 |
ENSDART00000173657
|
CU929416.1
|
ENSDARG00000105572 |
| chr12_-_4212010 | 0.34 |
ENSDART00000059298
|
zgc:92313
|
zgc:92313 |
| chr15_-_8786436 | 0.33 |
|
|
|
| chr13_-_8360779 | 0.32 |
ENSDART00000058107
|
mcfd2
|
multiple coagulation factor deficiency 2 |
| chr11_-_12024136 | 0.32 |
ENSDART00000111919
|
sp2
|
sp2 transcription factor |
| chr13_-_32324315 | 0.32 |
|
|
|
| chr13_-_33096839 | 0.32 |
ENSDART00000057386
|
golga5
|
golgin A5 |
| chr8_+_25940883 | 0.32 |
ENSDART00000140626
|
ENSDARG00000061023
|
ENSDARG00000061023 |
| chr7_-_19116856 | 0.31 |
ENSDART00000165680
|
ntn4
|
netrin 4 |
| chr4_-_5010148 | 0.31 |
ENSDART00000154025
|
strip2
|
striatin interacting protein 2 |
| chr17_-_14583728 | 0.30 |
ENSDART00000174703
|
daam1a
|
dishevelled associated activator of morphogenesis 1a |
| chr12_+_13704712 | 0.30 |
ENSDART00000152257
|
ppp1r16a
|
protein phosphatase 1, regulatory subunit 16A |
| chr23_-_27663219 | 0.30 |
ENSDART00000138381
|
si:ch211-156j22.4
|
si:ch211-156j22.4 |
| chr20_-_26492848 | 0.30 |
ENSDART00000078062
|
armt1
|
acidic residue methyltransferase 1 |
| chr5_-_36741219 | 0.30 |
ENSDART00000084675
|
wdr44
|
WD repeat domain 44 |
| chr14_+_29601073 | 0.30 |
ENSDART00000143763
|
fam149a
|
family with sequence similarity 149 member A |
| chr1_+_16683891 | 0.30 |
ENSDART00000103262
ENSDART00000145068 ENSDART00000169619 ENSDART00000010526 |
fat1a
|
FAT atypical cadherin 1a |
| chr5_-_31889551 | 0.30 |
ENSDART00000168870
|
gpr107
|
G protein-coupled receptor 107 |
| chr3_+_56882700 | 0.30 |
|
|
|
| chr6_+_38775771 | 0.29 |
ENSDART00000129655
|
ube3a
|
ubiquitin protein ligase E3A |
| chr11_+_30927094 | 0.29 |
ENSDART00000112098
|
stx10
|
syntaxin 10 |
| chr7_-_41446221 | 0.29 |
ENSDART00000099121
|
arl8
|
ADP-ribosylation factor-like 8 |
| chr5_+_50602106 | 0.29 |
ENSDART00000159571
|
msh3
|
mutS homolog 3 (E. coli) |
| chr8_-_11286377 | 0.29 |
ENSDART00000008215
|
pip5k1bb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta b |
| chr1_+_34741292 | 0.29 |
ENSDART00000109678
|
usp38
|
ubiquitin specific peptidase 38 |
| chr4_-_21745792 | 0.29 |
ENSDART00000066897
|
pawr
|
PRKC, apoptosis, WT1, regulator |
| chr19_+_7717962 | 0.28 |
ENSDART00000112404
|
cgnb
|
cingulin b |
| chr16_-_7003373 | 0.28 |
|
|
|
| chr16_+_32060521 | 0.28 |
ENSDART00000047570
|
mboat7
|
membrane bound O-acyltransferase domain containing 7 |
| chr8_+_31019730 | 0.28 |
|
|
|
| chr24_+_28304276 | 0.28 |
ENSDART00000018095
|
sh3glb1a
|
SH3-domain GRB2-like endophilin B1a |
| chr10_-_25366450 | 0.27 |
ENSDART00000123820
|
tmem135
|
transmembrane protein 135 |
| chr21_+_5854755 | 0.27 |
ENSDART00000149689
|
mob1bb
|
MOB kinase activator 1Bb |
| chr25_-_7794262 | 0.26 |
ENSDART00000156761
|
ambra1b
|
autophagy/beclin-1 regulator 1b |
| chr11_-_36212977 | 0.26 |
ENSDART00000165203
|
usp48
|
ubiquitin specific peptidase 48 |
| chr10_+_35313772 | 0.26 |
ENSDART00000028940
|
styxl1
|
serine/threonine/tyrosine interacting-like 1 |
| chr8_+_25072241 | 0.26 |
ENSDART00000143922
|
atxn7l2b
|
ataxin 7-like 2b |
| chr14_+_29601252 | 0.26 |
ENSDART00000143763
|
fam149a
|
family with sequence similarity 149 member A |
| chr22_-_6723168 | 0.25 |
|
|
|
| chr17_+_22291961 | 0.25 |
ENSDART00000151929
ENSDART00000089919 ENSDART00000000804 |
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr24_-_12685927 | 0.25 |
ENSDART00000039312
|
ipo4
|
importin 4 |
| chr13_-_30938624 | 0.25 |
ENSDART00000112653
|
wdfy4
|
WDFY family member 4 |
| chr12_+_13704914 | 0.25 |
ENSDART00000152257
|
ppp1r16a
|
protein phosphatase 1, regulatory subunit 16A |
| chr8_+_19589519 | 0.25 |
ENSDART00000144667
|
slc35a3a
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3a |
| chr20_-_34894930 | 0.24 |
|
|
|
| chr17_-_26916870 | 0.24 |
ENSDART00000139946
|
nipal3
|
NIPA-like domain containing 3 |
| chr23_+_23256859 | 0.24 |
ENSDART00000132296
|
klhl17
|
kelch-like family member 17 |
| chr8_+_52297522 | 0.23 |
ENSDART00000098439
|
ube2d4
|
ubiquitin-conjugating enzyme E2D 4 (putative) |
| chr11_-_26819525 | 0.23 |
ENSDART00000043091
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr9_-_38589607 | 0.23 |
ENSDART00000148268
|
znf142
|
zinc finger protein 142 |
| chr9_-_27600628 | 0.23 |
|
|
|
| chr22_-_6723094 | 0.23 |
|
|
|
| chr6_-_6895175 | 0.23 |
ENSDART00000149232
ENSDART00000150033 |
bin1b
|
bridging integrator 1b |
| chr5_-_29551051 | 0.23 |
ENSDART00000156048
|
zbtb44
|
zinc finger and BTB domain containing 44 |
| chr17_-_15659244 | 0.23 |
ENSDART00000142972
|
manea
|
mannosidase, endo-alpha |
| chr20_-_44020387 | 0.22 |
ENSDART00000026213
|
mark3b
|
MAP/microtubule affinity-regulating kinase 3b |
| chr20_-_34894607 | 0.22 |
|
|
|
| chr3_-_55219992 | 0.22 |
ENSDART00000175249
ENSDART00000153774 |
ern1
|
endoplasmic reticulum to nucleus signaling 1 |
| chr17_+_22291546 | 0.22 |
ENSDART00000151929
ENSDART00000089919 ENSDART00000000804 |
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr13_+_24157232 | 0.22 |
ENSDART00000134048
|
ccsapb
|
centriole, cilia and spindle-associated protein b |
| chr10_-_6587281 | 0.22 |
ENSDART00000163788
ENSDART00000171833 |
chd1
|
chromodomain helicase DNA binding protein 1 |
| chr3_-_36080647 | 0.22 |
ENSDART00000025326
|
csnk1da
|
casein kinase 1, delta a |
| chr18_+_18077901 | 0.21 |
|
|
|
| chr4_-_20514831 | 0.21 |
ENSDART00000146621
|
stk38l
|
serine/threonine kinase 38 like |
| chr1_+_39141680 | 0.21 |
ENSDART00000149984
|
irf2a
|
interferon regulatory factor 2a |
| chr8_+_53173077 | 0.21 |
ENSDART00000131514
|
nadka
|
NAD kinase a |
| chr10_-_6587375 | 0.21 |
ENSDART00000163788
ENSDART00000171833 |
chd1
|
chromodomain helicase DNA binding protein 1 |
| chr1_-_11191863 | 0.21 |
ENSDART00000163971
ENSDART00000123431 |
iqce
|
IQ motif containing E |
| chr1_-_39141284 | 0.21 |
ENSDART00000053763
|
dctd
|
dCMP deaminase |
| chr10_+_27000079 | 0.21 |
ENSDART00000089144
|
vps51
|
vacuolar protein sorting 51 homolog (S. cerevisiae) |
| chr6_+_19167599 | 0.20 |
ENSDART00000086619
|
prkca
|
protein kinase C, alpha |
| chr2_+_12140251 | 0.20 |
ENSDART00000124501
ENSDART00000153873 |
trove2
|
TROVE domain family, member 2 |
| chr12_-_35481361 | 0.20 |
ENSDART00000158658
ENSDART00000168958 ENSDART00000162175 |
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr1_-_18955249 | 0.20 |
ENSDART00000146688
|
polr1e
|
polymerase (RNA) I polypeptide E |
| chr3_+_15605866 | 0.20 |
ENSDART00000140160
|
BX914211.1
|
ENSDARG00000094116 |
| chr7_-_20378231 | 0.20 |
|
|
|
| chr8_-_41511321 | 0.20 |
ENSDART00000019858
|
golga1
|
golgin A1 |
| chr23_+_38335341 | 0.20 |
ENSDART00000177981
ENSDART00000178842 |
CABZ01070579.1
|
ENSDARG00000109193 |
| chr21_+_43183835 | 0.20 |
ENSDART00000175107
|
aff4
|
AF4/FMR2 family, member 4 |
| chr1_-_13740237 | 0.19 |
ENSDART00000123895
|
ENSDARG00000056677
|
ENSDARG00000056677 |
| chr8_-_11286437 | 0.19 |
ENSDART00000008215
|
pip5k1bb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta b |
| chr5_-_12586284 | 0.19 |
ENSDART00000051664
|
ypel1
|
yippee-like 1 |
| chr20_+_37592274 | 0.19 |
ENSDART00000153092
|
BX088686.1
|
ENSDARG00000096774 |
| chr19_-_35155287 | 0.19 |
ENSDART00000175621
|
elp2
|
elongator acetyltransferase complex subunit 2 |
| chr23_-_24616306 | 0.19 |
ENSDART00000088777
|
atp13a2
|
ATPase type 13A2 |
| chr2_+_19588085 | 0.19 |
ENSDART00000163875
|
cc2d1b
|
coiled-coil and C2 domain containing 1B |
| chr21_-_22399283 | 0.19 |
ENSDART00000137959
|
il7r
|
interleukin 7 receptor |
| chr16_+_32060609 | 0.19 |
ENSDART00000047570
|
mboat7
|
membrane bound O-acyltransferase domain containing 7 |
| chr11_-_2437361 | 0.19 |
|
|
|
| chr15_+_22510807 | 0.19 |
ENSDART00000040542
|
arhgef12a
|
Rho guanine nucleotide exchange factor (GEF) 12a |
| chr22_+_39098205 | 0.19 |
ENSDART00000002826
|
gmppb
|
GDP-mannose pyrophosphorylase B |
| chr5_+_32698516 | 0.19 |
ENSDART00000097945
|
usp20
|
ubiquitin specific peptidase 20 |
| chr22_+_23260413 | 0.18 |
|
|
|
| chr10_-_35313462 | 0.18 |
ENSDART00000139107
|
prr11
|
proline rich 11 |
| chr4_-_14193396 | 0.18 |
ENSDART00000101812
ENSDART00000143804 |
pus7l
|
pseudouridylate synthase 7-like |
| chr21_+_5854712 | 0.18 |
ENSDART00000149689
|
mob1bb
|
MOB kinase activator 1Bb |
| chr17_-_20110496 | 0.18 |
|
|
|
| chr3_-_20643874 | 0.18 |
ENSDART00000163473
ENSDART00000159457 |
spop
|
speckle-type POZ protein |
| chr15_+_31920039 | 0.18 |
|
|
|
| chr6_-_53391354 | 0.18 |
ENSDART00000155483
|
si:ch211-161c3.6
|
si:ch211-161c3.6 |
| chr12_-_22387786 | 0.18 |
ENSDART00000109707
|
neurl4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr16_+_37941736 | 0.18 |
ENSDART00000178753
|
cep162
|
centrosomal protein 162 |
| chr6_-_10574416 | 0.18 |
ENSDART00000104884
|
wipf1b
|
WAS/WASL interacting protein family, member 1b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:1902001 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) regulation of organic acid transport(GO:0032890) positive regulation of organic acid transport(GO:0032892) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transport(GO:1903793) regulation of anion transmembrane transport(GO:1903959) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.2 | 0.7 | GO:0051096 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.2 | 0.9 | GO:0015809 | arginine transport(GO:0015809) |
| 0.2 | 0.6 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.2 | 0.6 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.4 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.1 | 0.5 | GO:1903726 | negative regulation of phospholipid metabolic process(GO:1903726) |
| 0.1 | 0.5 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.1 | 0.4 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.7 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.1 | 1.2 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 0.5 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.1 | 1.2 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.1 | 0.3 | GO:0046950 | cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.1 | 0.6 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.5 | GO:0035264 | multicellular organism growth(GO:0035264) |
| 0.1 | 0.5 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.3 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 0.6 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.2 | GO:0046078 | dUMP biosynthetic process(GO:0006226) dUMP metabolic process(GO:0046078) |
| 0.1 | 0.2 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 0.3 | GO:1902019 | regulation of cilium beat frequency(GO:0003356) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 1.1 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.2 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.9 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.9 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.1 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.9 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.2 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.0 | 1.6 | GO:0007596 | blood coagulation(GO:0007596) hemostasis(GO:0007599) |
| 0.0 | 0.1 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.1 | GO:0051204 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.0 | 0.3 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 1.9 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.6 | GO:0000959 | mitochondrial RNA metabolic process(GO:0000959) |
| 0.0 | 0.2 | GO:0044241 | lipid digestion(GO:0044241) |
| 0.0 | 0.1 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 1.0 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.2 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.0 | 0.3 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.3 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.0 | 0.2 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.5 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.0 | 0.3 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0010660 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.0 | 0.2 | GO:0071108 | central nervous system morphogenesis(GO:0021551) protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.0 | 0.4 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 1.1 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.2 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.3 | GO:0021986 | habenula development(GO:0021986) |
| 0.0 | 0.4 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.4 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.1 | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis(GO:0090049) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.2 | 0.6 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.2 | 0.6 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.2 | 0.8 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.2 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 0.9 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.3 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.1 | 0.5 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 0.5 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.2 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 0.6 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 0.2 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 1.2 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 1.0 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.7 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.0 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) Sec62/Sec63 complex(GO:0031207) |
| 0.0 | 1.4 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.9 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 1.1 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0032135 | DNA insertion or deletion binding(GO:0032135) |
| 0.1 | 0.1 | GO:0035004 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) phosphatidylinositol 3-kinase activity(GO:0035004) |
| 0.1 | 0.6 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.6 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 0.7 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 0.8 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.2 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.1 | 0.3 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 0.9 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 1.2 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 1.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.6 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 2.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 1.8 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.3 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.6 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.1 | GO:0072572 | poly-ADP-D-ribose binding(GO:0072572) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.3 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.4 | GO:0071617 | lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.3 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.6 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 1.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.4 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.9 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 1.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.0 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 1.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.3 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.9 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.8 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |