Project
DANIO-CODE
Navigation
Downloads
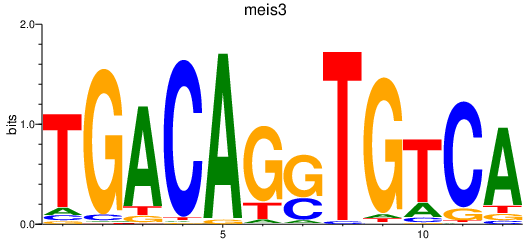
Results for meis3
Z-value: 0.70
Transcription factors associated with meis3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
meis3
|
ENSDARG00000002795 | myeloid ecotropic viral integration site 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| meis3 | dr10_dc_chr15_-_6866486_6866535 | 0.77 | 5.5e-04 | Click! |
Activity profile of meis3 motif
Sorted Z-values of meis3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of meis3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_26623251 | 2.22 |
ENSDART00000159264
|
sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr23_-_10242377 | 2.07 |
ENSDART00000129044
|
krt5
|
keratin 5 |
| chr22_-_15567180 | 1.74 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr7_-_26035308 | 1.58 |
ENSDART00000131906
|
zgc:77439
|
zgc:77439 |
| chr10_+_26705729 | 1.50 |
ENSDART00000147013
|
si:ch73-52f15.5
|
si:ch73-52f15.5 |
| chr8_-_7425677 | 1.29 |
ENSDART00000149671
|
hdac6
|
histone deacetylase 6 |
| chr23_+_21737142 | 1.20 |
ENSDART00000066125
|
dhrs3a
|
dehydrogenase/reductase (SDR family) member 3a |
| chr8_-_11191783 | 1.17 |
ENSDART00000131171
|
unc45b
|
unc-45 myosin chaperone B |
| chr2_+_45228446 | 1.16 |
ENSDART00000123966
|
chrng
|
cholinergic receptor, nicotinic, gamma |
| chr5_-_46382728 | 1.04 |
|
|
|
| chr7_+_40972616 | 0.99 |
ENSDART00000173814
|
scrib
|
scribbled planar cell polarity protein |
| chr1_-_40208544 | 0.96 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr1_-_40208469 | 0.95 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr8_+_53118919 | 0.92 |
|
|
|
| chr16_-_26663643 | 0.88 |
ENSDART00000008152
|
sgk2b
|
serum/glucocorticoid regulated kinase 2b |
| chr12_-_45714172 | 0.86 |
ENSDART00000149044
|
pax2b
|
paired box 2b |
| chr15_+_24064257 | 0.86 |
ENSDART00000156714
|
CR394545.1
|
ENSDARG00000097097 |
| chr16_-_16244210 | 0.83 |
ENSDART00000012718
|
fabp11b
|
fatty acid binding protein 11b |
| chr14_-_26138828 | 0.83 |
ENSDART00000140173
|
si:dkeyp-110e4.6
|
si:dkeyp-110e4.6 |
| chr20_-_53560663 | 0.80 |
ENSDART00000146001
|
wasf1
|
WAS protein family, member 1 |
| chr23_-_10242235 | 0.78 |
ENSDART00000081223
ENSDART00000144280 |
krt5
|
keratin 5 |
| chr17_-_15659244 | 0.77 |
ENSDART00000142972
|
manea
|
mannosidase, endo-alpha |
| chr1_+_1657493 | 0.75 |
ENSDART00000103850
|
atp1a1a.3
|
ATPase, Na+/K+ transporting, alpha 1a polypeptide, tandem duplicate 3 |
| chr10_+_404464 | 0.70 |
ENSDART00000145124
ENSDART00000124319 |
dact3a
|
dishevelled-binding antagonist of beta-catenin 3a |
| chr7_-_26035541 | 0.70 |
ENSDART00000057288
|
zgc:77439
|
zgc:77439 |
| chr20_-_26568204 | 0.69 |
ENSDART00000158213
|
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr22_-_10372023 | 0.68 |
|
|
|
| chr15_-_25430003 | 0.66 |
ENSDART00000152651
|
cluha
|
clustered mitochondria (cluA/CLU1) homolog a |
| chr8_+_8252892 | 0.66 |
ENSDART00000170566
|
srpk3
|
SRSF protein kinase 3 |
| chr15_-_34198725 | 0.62 |
ENSDART00000170130
|
ENSDARG00000101697
|
ENSDARG00000101697 |
| chr3_+_55779906 | 0.60 |
|
|
|
| chr2_-_44330405 | 0.60 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
| chr21_-_45573799 | 0.59 |
ENSDART00000158489
|
zgc:77058
|
zgc:77058 |
| chr23_-_38565787 | 0.58 |
|
|
|
| chr15_-_34198695 | 0.57 |
ENSDART00000170130
|
ENSDARG00000101697
|
ENSDARG00000101697 |
| chr22_+_10372455 | 0.57 |
ENSDART00000122403
|
wu:fb55g09
|
wu:fb55g09 |
| chr14_-_6624878 | 0.56 |
ENSDART00000166439
|
si:ch211-266k2.1
|
si:ch211-266k2.1 |
| chr2_-_32575527 | 0.55 |
ENSDART00000140026
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr23_-_10241885 | 0.54 |
ENSDART00000081223
ENSDART00000144280 |
krt5
|
keratin 5 |
| chr18_-_19114558 | 0.54 |
ENSDART00000177621
|
dennd4a
|
DENN/MADD domain containing 4A |
| chr7_-_18629056 | 0.51 |
ENSDART00000021502
|
mllt3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr2_+_45843528 | 0.51 |
ENSDART00000114225
ENSDART00000169279 |
CU467828.1
|
ENSDARG00000074082 |
| KN150670v1_-_61400 | 0.47 |
|
|
|
| chr8_-_27496134 | 0.47 |
ENSDART00000134879
|
si:ch211-254n4.3
|
si:ch211-254n4.3 |
| chr23_-_38565708 | 0.46 |
|
|
|
| chr22_-_22206778 | 0.44 |
ENSDART00000105580
|
ap3d1
|
adaptor-related protein complex 3, delta 1 subunit |
| chr9_-_28588288 | 0.42 |
ENSDART00000104317
ENSDART00000064343 |
klf7b
|
Kruppel-like factor 7b |
| chr20_+_51291139 | 0.42 |
ENSDART00000073981
|
eif2s1b
|
eukaryotic translation initiation factor 2, subunit 1 alpha b |
| chr8_-_19166630 | 0.39 |
|
|
|
| chr5_+_23741791 | 0.39 |
ENSDART00000049003
|
atp6v1aa
|
ATPase, H+ transporting, lysosomal, V1 subunit Aa |
| chr13_+_22165657 | 0.39 |
|
|
|
| chr20_-_23272102 | 0.37 |
ENSDART00000167197
|
spata18
|
spermatogenesis associated 18 |
| chr14_-_17000025 | 0.37 |
ENSDART00000168853
|
jakmip1
|
janus kinase and microtubule interacting protein 1 |
| chr5_+_22613963 | 0.35 |
ENSDART00000128271
|
NEXMIF (1 of many)
|
si:dkey-114c15.7 |
| chr16_-_7321943 | 0.35 |
ENSDART00000149260
|
nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| chr13_+_22165699 | 0.33 |
|
|
|
| chr11_-_17846665 | 0.33 |
ENSDART00000005999
|
twf2b
|
twinfilin actin-binding protein 2b |
| chr25_+_32114076 | 0.31 |
ENSDART00000156145
|
scaper
|
S-phase cyclin A-associated protein in the ER |
| chr22_+_21095535 | 0.31 |
|
|
|
| chr16_-_26663608 | 0.27 |
ENSDART00000134908
|
sgk2b
|
serum/glucocorticoid regulated kinase 2b |
| chr11_-_27970634 | 0.27 |
|
|
|
| chr6_+_33552572 | 0.27 |
ENSDART00000040334
|
pik3r3b
|
phosphoinositide-3-kinase, regulatory subunit 3b (gamma) |
| chr19_-_36009321 | 0.27 |
ENSDART00000136811
|
col8a2
|
collagen, type VIII, alpha 2 |
| chr16_-_12095302 | 0.27 |
ENSDART00000148666
ENSDART00000029121 |
usp5
|
ubiquitin specific peptidase 5 (isopeptidase T) |
| chr9_-_34410053 | 0.25 |
ENSDART00000111027
|
dcaf6
|
ddb1 and cul4 associated factor 6 |
| chr8_-_4451417 | 0.25 |
ENSDART00000141915
|
si:ch211-166a6.5
|
si:ch211-166a6.5 |
| chr4_+_27139902 | 0.24 |
|
|
|
| chr3_+_44468134 | 0.24 |
ENSDART00000164561
|
ENSDARG00000098547
|
ENSDARG00000098547 |
| chr7_-_71929355 | 0.24 |
|
|
|
| chr20_-_26521700 | 0.24 |
ENSDART00000110883
|
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr9_+_50341397 | 0.22 |
|
|
|
| chr4_+_21091640 | 0.21 |
ENSDART00000005847
|
nav3
|
neuron navigator 3 |
| chr11_+_2437479 | 0.20 |
ENSDART00000130886
|
NABP2
|
nucleic acid binding protein 2 |
| chr23_+_23255489 | 0.19 |
ENSDART00000137353
|
klhl17
|
kelch-like family member 17 |
| chr6_+_15635696 | 0.19 |
ENSDART00000128939
|
iqca1
|
IQ motif containing with AAA domain 1 |
| chr9_-_25370657 | 0.18 |
ENSDART00000166684
|
esd
|
esterase D/formylglutathione hydrolase |
| chr6_+_15635738 | 0.17 |
ENSDART00000128939
|
iqca1
|
IQ motif containing with AAA domain 1 |
| chr2_+_26824005 | 0.15 |
ENSDART00000172669
ENSDART00000056795 ENSDART00000144837 |
hectd3
|
HECT domain containing 3 |
| chr21_+_10693199 | 0.15 |
|
|
|
| chr20_+_46969915 | 0.14 |
ENSDART00000158124
|
ENSDARG00000103380
|
ENSDARG00000103380 |
| chr24_+_12845250 | 0.09 |
ENSDART00000126842
|
flj11011l
|
hypothetical protein FLJ11011-like (H. sapiens) |
| chr3_+_26603352 | 0.08 |
ENSDART00000114552
|
si:dkey-202l16.5
|
si:dkey-202l16.5 |
| chr4_-_17033790 | 0.08 |
|
|
|
| chr7_-_18629119 | 0.07 |
ENSDART00000021502
|
mllt3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr17_-_11264425 | 0.07 |
ENSDART00000151847
|
arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr6_+_8871439 | 0.07 |
ENSDART00000083820
|
vps16
|
vacuolar protein sorting protein 16 |
| chr23_-_33753634 | 0.07 |
ENSDART00000138416
|
tfcp2
|
transcription factor CP2 |
| chr9_-_25370467 | 0.04 |
ENSDART00000141837
|
esd
|
esterase D/formylglutathione hydrolase |
| chr7_-_33791033 | 0.04 |
ENSDART00000052404
|
map2k5
|
mitogen-activated protein kinase kinase 5 |
| chr8_-_41511380 | 0.00 |
ENSDART00000019858
|
golga1
|
golgin A1 |
| chr19_-_35155287 | 0.00 |
ENSDART00000175621
|
elp2
|
elongator acetyltransferase complex subunit 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.2 | 1.0 | GO:1902946 | protein localization to early endosome(GO:1902946) |
| 0.1 | 1.9 | GO:0030656 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.1 | 0.4 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.1 | 0.5 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 1.2 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 1.2 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 0.4 | GO:1904969 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.1 | 0.8 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.6 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 0.3 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.6 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.2 | GO:0046292 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.0 | 2.2 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 3.4 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 1.2 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 2.3 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.5 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 1.3 | GO:0006476 | protein deacetylation(GO:0006476) |
| 0.0 | 0.7 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.2 | GO:0072576 | liver morphogenesis(GO:0072576) |
| 0.0 | 0.1 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.4 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.1 | 0.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.2 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 1.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.4 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.4 | 1.3 | GO:0047611 | acetylspermidine deacetylase activity(GO:0047611) |
| 0.2 | 1.2 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.2 | 2.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.7 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.2 | GO:0018738 | S-formylglutathione hydrolase activity(GO:0018738) |
| 0.1 | 1.1 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.8 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.4 | GO:0043022 | ribosome binding(GO:0043022) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 2.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.3 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | REACTOME ACETYLCHOLINE BINDING AND DOWNSTREAM EVENTS | Genes involved in Acetylcholine Binding And Downstream Events |
| 0.0 | 1.3 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.8 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |