Project
DANIO-CODE
Navigation
Downloads
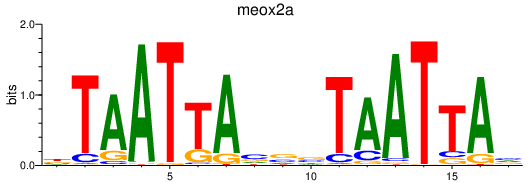
Results for meox2a
Z-value: 0.42
Transcription factors associated with meox2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
meox2a
|
ENSDARG00000040911 | mesenchyme homeobox 2a |
Activity profile of meox2a motif
Sorted Z-values of meox2a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of meox2a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_28559915 | 1.14 |
ENSDART00000088827
|
dpp7
|
dipeptidyl-peptidase 7 |
| chr19_-_43037791 | 0.83 |
ENSDART00000004392
|
fkbp9
|
FK506 binding protein 9 |
| chr6_+_55022668 | 0.69 |
ENSDART00000158845
|
mybphb
|
myosin binding protein Hb |
| chr17_+_17935213 | 0.63 |
ENSDART00000104999
|
ccdc85ca
|
coiled-coil domain containing 85C, a |
| chr5_+_48031920 | 0.63 |
ENSDART00000008043
ENSDART00000171438 |
adgrv1
|
adhesion G protein-coupled receptor V1 |
| chr20_-_22576513 | 0.56 |
ENSDART00000103510
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr10_+_7125001 | 0.55 |
ENSDART00000157987
|
psd3l
|
pleckstrin and Sec7 domain containing 3, like |
| chr4_+_9668755 | 0.54 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr19_-_32562731 | 0.54 |
ENSDART00000151407
|
pag1
|
phosphoprotein membrane anchor with glycosphingolipid microdomains 1 |
| chr23_+_36007936 | 0.53 |
ENSDART00000128533
|
hoxc3a
|
homeo box C3a |
| chr19_+_12995955 | 0.52 |
ENSDART00000132892
|
cthrc1a
|
collagen triple helix repeat containing 1a |
| chr11_-_29910947 | 0.52 |
ENSDART00000156121
|
scml2
|
sex comb on midleg-like 2 (Drosophila) |
| chr3_-_15584548 | 0.49 |
ENSDART00000137325
|
mvp
|
major vault protein |
| chr6_-_22504772 | 0.48 |
ENSDART00000170039
|
sept9b
|
septin 9b |
| chr11_-_41219340 | 0.48 |
ENSDART00000109204
|
megf6b
|
multiple EGF-like-domains 6b |
| chr19_-_43038013 | 0.47 |
ENSDART00000004392
|
fkbp9
|
FK506 binding protein 9 |
| chr21_-_5852663 | 0.47 |
ENSDART00000130521
|
CACFD1
|
calcium channel flower domain containing 1 |
| chr11_+_8558222 | 0.47 |
ENSDART00000169141
ENSDART00000126523 |
tbl1xr1a
|
transducin (beta)-like 1 X-linked receptor 1a |
| chr2_-_23348612 | 0.44 |
ENSDART00000110373
|
znf414
|
zinc finger protein 414 |
| chr12_+_13080209 | 0.43 |
ENSDART00000127870
|
cmn
|
calymmin |
| chr16_-_43441084 | 0.43 |
ENSDART00000058680
|
psma2
|
proteasome subunit alpha 2 |
| chr23_-_23474703 | 0.42 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr16_+_24045774 | 0.42 |
ENSDART00000133484
|
apoeb
|
apolipoprotein Eb |
| chr3_+_36282205 | 0.41 |
ENSDART00000170318
|
si:ch1073-443f11.2
|
si:ch1073-443f11.2 |
| chr22_-_10430130 | 0.41 |
ENSDART00000064801
|
ogn
|
osteoglycin |
| chr4_-_18606513 | 0.40 |
ENSDART00000049061
|
cdkn1ba
|
cyclin-dependent kinase inhibitor 1Ba |
| chr24_+_38371710 | 0.39 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr12_+_30471781 | 0.39 |
ENSDART00000126984
|
nrap
|
nebulin-related anchoring protein |
| chr12_-_36565562 | 0.39 |
ENSDART00000153259
|
si:ch211-216b21.2
|
si:ch211-216b21.2 |
| chr2_+_33399405 | 0.39 |
ENSDART00000137207
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| KN149998v1_+_54953 | 0.38 |
|
|
|
| chr16_+_54906517 | 0.38 |
ENSDART00000126646
|
ENSDARG00000091079
|
ENSDARG00000091079 |
| chr24_-_1155215 | 0.37 |
ENSDART00000177356
|
itgb1a
|
integrin, beta 1a |
| chr13_+_19191645 | 0.37 |
ENSDART00000058036
|
emx2
|
empty spiracles homeobox 2 |
| chr14_+_21816442 | 0.37 |
ENSDART00000109759
|
tmx2a
|
thioredoxin-related transmembrane protein 2a |
| chr14_+_6122948 | 0.37 |
ENSDART00000149783
ENSDART00000148461 |
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr24_-_31772736 | 0.37 |
|
|
|
| chr10_-_8101868 | 0.36 |
ENSDART00000179549
|
si:ch211-251f6.6
|
si:ch211-251f6.6 |
| chr6_-_14162846 | 0.36 |
ENSDART00000100762
|
inpp4ab
|
inositol polyphosphate-4-phosphatase type I Ab |
| chr25_+_33550125 | 0.36 |
ENSDART00000153934
|
CT573383.1
|
ENSDARG00000097513 |
| chr3_+_51837866 | 0.35 |
ENSDART00000104650
|
ENSDARG00000074362
|
ENSDARG00000074362 |
| chr12_+_28739504 | 0.35 |
ENSDART00000152991
|
nfe2l1b
|
nuclear factor, erythroid 2-like 1b |
| KN150207v1_-_1208 | 0.35 |
ENSDART00000171112
|
CABZ01112575.1
|
ENSDARG00000099354 |
| chr14_+_25168063 | 0.35 |
ENSDART00000173436
|
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr16_-_9108885 | 0.35 |
ENSDART00000153785
|
triob
|
trio Rho guanine nucleotide exchange factor b |
| chr10_+_9052152 | 0.35 |
ENSDART00000139466
|
itga2.2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor), tandem duplicate 2 |
| chr9_+_30774173 | 0.35 |
ENSDART00000160590
|
tbc1d4
|
TBC1 domain family, member 4 |
| chr25_+_19992389 | 0.35 |
ENSDART00000143441
|
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr5_-_62816208 | 0.34 |
ENSDART00000097325
|
c5
|
complement component 5 |
| chr12_+_17382681 | 0.33 |
ENSDART00000020628
|
cyth3a
|
cytohesin 3a |
| chr16_+_17061069 | 0.33 |
ENSDART00000111074
|
si:ch211-120k19.1
|
si:ch211-120k19.1 |
| chr21_+_26035166 | 0.33 |
ENSDART00000134939
|
rpl23a
|
ribosomal protein L23a |
| chr13_-_1278448 | 0.33 |
ENSDART00000049684
|
bag2
|
BCL2-associated athanogene 2 |
| chr11_-_21422588 | 0.33 |
|
|
|
| chr21_-_26035057 | 0.32 |
ENSDART00000141382
|
rab34b
|
RAB34, member RAS oncogene family b |
| chr14_+_49921861 | 0.32 |
ENSDART00000173240
|
zgc:154054
|
zgc:154054 |
| chr16_+_43441389 | 0.32 |
ENSDART00000085282
|
mrpl32
|
mitochondrial ribosomal protein L32 |
| chr10_+_31362144 | 0.32 |
|
|
|
| chr1_-_23771836 | 0.32 |
ENSDART00000126950
|
sh3d19
|
SH3 domain containing 19 |
| chr12_+_13614102 | 0.32 |
ENSDART00000152689
|
oplah
|
5-oxoprolinase (ATP-hydrolysing) |
| chr16_+_14697969 | 0.31 |
ENSDART00000133566
|
deptor
|
DEP domain containing MTOR-interacting protein |
| chr2_+_35871754 | 0.31 |
ENSDART00000134918
|
dhx9
|
DEAH (Asp-Glu-Ala-His) box helicase 9 |
| chr4_-_22590638 | 0.31 |
ENSDART00000137814
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr21_-_43020159 | 0.31 |
ENSDART00000065097
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr22_-_37417903 | 0.31 |
ENSDART00000149948
|
FP102167.1
|
ENSDARG00000095844 |
| chr14_+_31410970 | 0.30 |
ENSDART00000169796
|
si:dkeyp-11e3.1
|
si:dkeyp-11e3.1 |
| chr6_-_54816183 | 0.30 |
ENSDART00000148462
|
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr17_-_49878964 | 0.30 |
ENSDART00000154728
|
col12a1a
|
collagen, type XII, alpha 1a |
| chr3_+_22412059 | 0.30 |
|
|
|
| chr10_-_20567013 | 0.30 |
ENSDART00000159060
|
ddhd2
|
DDHD domain containing 2 |
| chr3_+_24327586 | 0.29 |
ENSDART00000153551
|
cbx6b
|
chromobox homolog 6b |
| chr2_-_17564676 | 0.29 |
ENSDART00000144251
|
artnb
|
artemin b |
| chr9_+_4410206 | 0.29 |
|
|
|
| chr23_+_44137593 | 0.29 |
ENSDART00000148470
|
CU993818.1
|
ENSDARG00000095873 |
| chr4_-_75641806 | 0.29 |
ENSDART00000174383
|
psmb10
|
proteasome subunit beta 10 |
| chr23_-_45927723 | 0.29 |
|
|
|
| chr9_-_1701626 | 0.29 |
ENSDART00000163482
|
hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr8_+_14848685 | 0.28 |
ENSDART00000146589
|
soat1
|
sterol O-acyltransferase 1 |
| chr7_+_26491774 | 0.27 |
ENSDART00000167956
|
tspan18a
|
tetraspanin 18a |
| chr20_-_3970778 | 0.27 |
ENSDART00000178724
ENSDART00000178565 |
TRIM67
|
tripartite motif containing 67 |
| chr8_-_46313975 | 0.27 |
ENSDART00000075189
|
mtor
|
mechanistic target of rapamycin (serine/threonine kinase) |
| chr3_-_40159349 | 0.27 |
ENSDART00000055186
|
atp5j2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F2 |
| chr17_-_12812747 | 0.27 |
ENSDART00000022874
ENSDART00000046025 |
psma6a
|
proteasome subunit alpha 6a |
| chr5_+_62611688 | 0.27 |
ENSDART00000177108
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr6_-_23193752 | 0.27 |
ENSDART00000159749
|
usp43a
|
ubiquitin specific peptidase 43a |
| chr21_-_11561855 | 0.27 |
ENSDART00000162426
|
cast
|
calpastatin |
| chr5_+_53178362 | 0.27 |
ENSDART00000169565
|
sat2b
|
spermidine/spermine N1-acetyltransferase family member 2b |
| chr22_-_26971466 | 0.27 |
ENSDART00000087202
|
ENSDARG00000061256
|
ENSDARG00000061256 |
| chr12_-_37124892 | 0.26 |
ENSDART00000146142
|
pmp22b
|
peripheral myelin protein 22b |
| chr5_+_50379968 | 0.26 |
ENSDART00000050988
|
gcnt4a
|
glucosaminyl (N-acetyl) transferase 4, core 2, a |
| chr11_-_204974 | 0.26 |
ENSDART00000173151
|
tegt
|
testis enhanced gene transcript (BAX inhibitor 1) |
| chr1_-_35525344 | 0.26 |
|
|
|
| chr15_-_18130992 | 0.26 |
ENSDART00000113142
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr8_+_52633069 | 0.26 |
ENSDART00000162953
|
cyr61l2
|
cysteine-rich, angiogenic inducer, 61 like 2 |
| chr12_-_1544844 | 0.25 |
|
|
|
| chr14_+_36529003 | 0.25 |
ENSDART00000111674
|
F2RL3
|
F2R like thrombin or trypsin receptor 3 |
| chr10_+_9259213 | 0.25 |
ENSDART00000064966
|
snx18b
|
sorting nexin 18b |
| chr10_-_44441481 | 0.25 |
ENSDART00000160231
|
sbno1
|
strawberry notch homolog 1 (Drosophila) |
| chr5_+_2384929 | 0.25 |
|
|
|
| chr21_+_28921734 | 0.25 |
ENSDART00000166575
|
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr12_-_48493654 | 0.24 |
ENSDART00000162603
|
ndufb8
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8 |
| chr19_+_14197118 | 0.24 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr25_+_5922190 | 0.24 |
ENSDART00000128816
|
sv2
|
synaptic vesicle glycoprotein 2 |
| chr4_+_377829 | 0.24 |
ENSDART00000030215
|
rpl18a
|
ribosomal protein L18a |
| chr11_-_1792616 | 0.24 |
|
|
|
| chr5_-_71340996 | 0.24 |
ENSDART00000162526
|
smyd1a
|
SET and MYND domain containing 1a |
| chr9_-_55204516 | 0.24 |
|
|
|
| chr21_-_5774127 | 0.24 |
ENSDART00000009241
|
rpl35
|
ribosomal protein L35 |
| chr7_-_71566426 | 0.23 |
ENSDART00000166724
|
myom1b
|
myomesin 1b |
| chr20_-_11179880 | 0.23 |
ENSDART00000152246
|
flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr22_-_864745 | 0.23 |
ENSDART00000035514
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr14_+_33073643 | 0.23 |
ENSDART00000052780
|
ndufa1
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1 |
| chr3_+_36155364 | 0.23 |
ENSDART00000059533
|
wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr17_+_25500291 | 0.23 |
ENSDART00000041721
|
aim1a
|
crystallin beta-gamma domain containing 1a |
| chr3_+_28822408 | 0.22 |
ENSDART00000133528
|
lgals2a
|
lectin, galactoside-binding, soluble, 2a |
| chr24_+_24686591 | 0.22 |
ENSDART00000080969
|
trim55b
|
tripartite motif containing 55b |
| chr4_-_11751943 | 0.22 |
ENSDART00000102301
|
podxl
|
podocalyxin-like |
| chr7_-_38288929 | 0.22 |
|
|
|
| chr4_+_149696 | 0.22 |
ENSDART00000161055
|
dusp16
|
dual specificity phosphatase 16 |
| chr13_+_4239960 | 0.22 |
ENSDART00000135113
|
sft2d1
|
SFT2 domain containing 1 |
| chr10_-_35293024 | 0.21 |
ENSDART00000145804
|
ypel2a
|
yippee-like 2a |
| chr20_-_47680305 | 0.21 |
ENSDART00000023058
|
efhc1
|
EF-hand domain (C-terminal) containing 1 |
| chr18_+_16997001 | 0.21 |
ENSDART00000147377
|
si:ch211-218c6.8
|
si:ch211-218c6.8 |
| chr25_-_16658906 | 0.21 |
ENSDART00000124729
ENSDART00000110859 |
ribc2
|
RIB43A domain with coiled-coils 2 |
| chr4_-_8005840 | 0.21 |
ENSDART00000036153
|
ccdc3a
|
coiled-coil domain containing 3a |
| chr13_-_40190349 | 0.21 |
ENSDART00000009343
|
pyroxd2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr23_+_45803725 | 0.21 |
ENSDART00000135602
|
si:ch73-290k24.6
|
si:ch73-290k24.6 |
| chr2_-_25306278 | 0.21 |
ENSDART00000132050
|
hltf
|
helicase-like transcription factor |
| chr14_-_11150618 | 0.21 |
ENSDART00000110424
|
si:ch211-153b23.4
|
si:ch211-153b23.4 |
| chr8_+_49789789 | 0.20 |
ENSDART00000083790
|
ntrk2a
|
neurotrophic tyrosine kinase, receptor, type 2a |
| chr23_-_24790203 | 0.20 |
|
|
|
| chr5_+_42406222 | 0.20 |
ENSDART00000009182
|
aqp3a
|
aquaporin 3a |
| chr19_-_16126342 | 0.20 |
|
|
|
| chr3_+_23538277 | 0.19 |
ENSDART00000111227
|
hoxb10a
|
homeo box B10a |
| chr16_+_5284778 | 0.19 |
ENSDART00000156685
ENSDART00000156765 |
soga3a
soga3a
|
SOGA family member 3a SOGA family member 3a |
| chr18_+_17594381 | 0.19 |
ENSDART00000010998
|
slc12a3
|
solute carrier family 12 (sodium/chloride transporter), member 3 |
| chr23_-_18203680 | 0.19 |
ENSDART00000016976
|
nucks1b
|
nuclear casein kinase and cyclin-dependent kinase substrate 1b |
| chr2_-_17721575 | 0.19 |
ENSDART00000141188
ENSDART00000100201 |
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr15_-_2224784 | 0.19 |
ENSDART00000009564
|
shox2
|
short stature homeobox 2 |
| chr8_-_13934852 | 0.19 |
ENSDART00000133830
|
serping1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr3_+_25888520 | 0.19 |
ENSDART00000135389
|
foxred2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr5_-_66792947 | 0.19 |
ENSDART00000147009
|
si:dkey-251i10.2
|
si:dkey-251i10.2 |
| chr1_+_16900546 | 0.19 |
ENSDART00000078889
|
helt
|
helt bHLH transcription factor |
| chr9_-_25437880 | 0.18 |
|
|
|
| chr25_-_3633631 | 0.18 |
ENSDART00000159335
ENSDART00000088077 |
zgc:158398
|
zgc:158398 |
| chr4_+_73490746 | 0.18 |
ENSDART00000174082
|
nup50
|
nucleoporin 50 |
| chr10_+_40239012 | 0.18 |
|
|
|
| chr13_+_9216834 | 0.18 |
ENSDART00000122695
|
pomk
|
protein-O-mannose kinase |
| chr19_-_28273791 | 0.18 |
ENSDART00000137033
|
srd5a1
|
steroid-5-alpha-reductase, alpha polypeptide 1 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 1) |
| chr18_+_5260471 | 0.18 |
ENSDART00000150992
|
wdr59
|
WD repeat domain 59 |
| chr10_-_22065636 | 0.18 |
|
|
|
| chr7_-_9557990 | 0.18 |
ENSDART00000055593
|
aldh1a3
|
aldehyde dehydrogenase 1 family, member A3 |
| chr25_+_4918339 | 0.17 |
ENSDART00000153980
|
parvb
|
parvin, beta |
| chr5_+_27897765 | 0.17 |
ENSDART00000166737
ENSDART00000164418 |
nfr
|
notochord formation related |
| chr10_+_8897426 | 0.17 |
ENSDART00000145596
|
itga1
|
integrin, alpha 1 |
| chr21_-_9313788 | 0.17 |
|
|
|
| chr12_+_30471689 | 0.17 |
ENSDART00000124920
|
nrap
|
nebulin-related anchoring protein |
| chr24_+_21395671 | 0.17 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr7_+_66660781 | 0.17 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr2_+_57687162 | 0.17 |
ENSDART00000147966
|
si:dkeyp-68b7.10
|
si:dkeyp-68b7.10 |
| chr11_+_25275546 | 0.16 |
|
|
|
| chr3_-_31392769 | 0.16 |
|
|
|
| chr12_+_46689953 | 0.16 |
ENSDART00000153035
|
tvp23b
|
trans-golgi network vesicle protein 23 homolog B (S. cerevisiae) |
| chr5_-_23649439 | 0.16 |
|
|
|
| chr23_-_27645138 | 0.16 |
ENSDART00000008174
|
pfkma
|
phosphofructokinase, muscle a |
| chr12_+_18559530 | 0.16 |
ENSDART00000152948
|
rgs9b
|
regulator of G protein signaling 9b |
| chr11_+_22213887 | 0.16 |
ENSDART00000174683
|
ppfia4
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 |
| chr7_-_51186389 | 0.16 |
ENSDART00000174328
|
arhgap6
|
Rho GTPase activating protein 6 |
| chr17_+_16415792 | 0.16 |
ENSDART00000064233
|
efcab11
|
EF-hand calcium binding domain 11 |
| chr10_-_11303185 | 0.15 |
ENSDART00000146727
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr19_+_14197020 | 0.15 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr7_+_69664384 | 0.15 |
ENSDART00000058763
|
sec24d
|
SEC24 homolog D, COPII coat complex component |
| chr19_+_20164849 | 0.15 |
ENSDART00000169017
|
hoxa11a
|
homeobox A11a |
| chr3_-_45420882 | 0.15 |
ENSDART00000161507
|
zgc:153426
|
zgc:153426 |
| chr21_+_13196717 | 0.15 |
ENSDART00000163767
|
adora2ab
|
adenosine A2a receptor b |
| chr16_+_54350976 | 0.15 |
ENSDART00000172622
ENSDART00000020033 |
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr2_+_1055716 | 0.15 |
ENSDART00000165477
|
AL935300.3
|
ENSDARG00000098661 |
| chr13_-_3801122 | 0.15 |
|
|
|
| chr3_-_13771322 | 0.15 |
ENSDART00000159177
ENSDART00000165174 |
si:dkey-61n16.5
|
si:dkey-61n16.5 |
| chr21_-_2116499 | 0.15 |
ENSDART00000167307
|
AL627305.3
|
ENSDARG00000102019 |
| chr8_-_51859979 | 0.14 |
|
|
|
| chr10_-_13281047 | 0.14 |
ENSDART00000001253
|
si:busm1-57f23.1
|
si:busm1-57f23.1 |
| chr25_+_29631901 | 0.14 |
ENSDART00000154849
|
ENSDARG00000029431
|
ENSDARG00000029431 |
| chr8_+_44765658 | 0.14 |
ENSDART00000025875
|
ENSDARG00000006901
|
ENSDARG00000006901 |
| chr11_-_25181234 | 0.14 |
ENSDART00000013714
|
gata1a
|
GATA binding protein 1a |
| chr25_+_13635079 | 0.14 |
ENSDART00000161012
|
ccdc135
|
coiled-coil domain containing 135 |
| chr15_+_34007335 | 0.14 |
ENSDART00000166141
|
tekt1
|
tektin 1 |
| chr17_+_23975487 | 0.14 |
|
|
|
| chr13_+_45843396 | 0.14 |
ENSDART00000005195
ENSDART00000074547 |
bsdc1
|
BSD domain containing 1 |
| chr6_-_17699493 | 0.13 |
ENSDART00000154180
|
CR388029.1
|
ENSDARG00000097214 |
| chr25_-_2386615 | 0.13 |
|
|
|
| chr5_+_23741791 | 0.13 |
ENSDART00000049003
|
atp6v1aa
|
ATPase, H+ transporting, lysosomal, V1 subunit Aa |
| chr25_-_6096124 | 0.13 |
ENSDART00000067512
|
psma4
|
proteasome subunit alpha 4 |
| chr4_+_13569588 | 0.13 |
ENSDART00000136152
|
calua
|
calumenin a |
| chr1_-_25369419 | 0.13 |
ENSDART00000168640
|
pdcd4a
|
programmed cell death 4a |
| chr19_+_21781738 | 0.13 |
|
|
|
| chr8_+_49076378 | 0.13 |
ENSDART00000032277
|
ddx51
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 |
| chr7_+_7304079 | 0.13 |
|
|
|
| chr3_+_13487523 | 0.13 |
ENSDART00000166000
|
si:ch211-194b1.1
|
si:ch211-194b1.1 |
| chr23_-_33017920 | 0.13 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0030818 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclic nucleotide metabolic process(GO:0030800) negative regulation of cyclic nucleotide biosynthetic process(GO:0030803) negative regulation of nucleotide biosynthetic process(GO:0030809) negative regulation of cAMP metabolic process(GO:0030815) negative regulation of cAMP biosynthetic process(GO:0030818) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) negative regulation of purine nucleotide biosynthetic process(GO:1900372) |
| 0.2 | 0.7 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.1 | 0.3 | GO:0071831 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.1 | 0.4 | GO:0099565 | chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.1 | 0.3 | GO:0034434 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.1 | 0.3 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 0.1 | GO:0035676 | anterior lateral line neuromast hair cell development(GO:0035676) |
| 0.1 | 0.2 | GO:0033628 | regulation of cell adhesion mediated by integrin(GO:0033628) positive regulation of cell adhesion mediated by integrin(GO:0033630) cell-cell adhesion mediated by integrin(GO:0033631) regulation of cell-cell adhesion mediated by integrin(GO:0033632) positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.3 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.1 | 0.2 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.1 | 0.3 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.4 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.1 | 0.2 | GO:0002526 | acute inflammatory response(GO:0002526) |
| 0.1 | 0.6 | GO:0003315 | heart rudiment formation(GO:0003315) |
| 0.1 | 0.3 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 0.4 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.1 | 0.3 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 0.2 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.1 | 0.2 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.4 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 1.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.9 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.4 | GO:0060118 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.0 | 0.5 | GO:1903038 | negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.2 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.1 | GO:0032475 | otolith formation(GO:0032475) |
| 0.0 | 0.2 | GO:0036268 | swimming(GO:0036268) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.4 | GO:1903672 | positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.1 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.7 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.8 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.0 | 0.1 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.2 | GO:0006735 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.1 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.0 | 0.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.2 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.0 | 0.1 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.0 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.1 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:1902868 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.0 | 0.1 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.6 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.3 | GO:0006749 | glutathione metabolic process(GO:0006749) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0032420 | photoreceptor inner segment(GO:0001917) stereocilium(GO:0032420) |
| 0.1 | 0.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 0.8 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.5 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.5 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.6 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 0.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.4 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.4 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 0.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.2 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.1 | 0.3 | GO:0019809 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.1 | 0.2 | GO:0015168 | polyol transmembrane transporter activity(GO:0015166) glycerol transmembrane transporter activity(GO:0015168) |
| 0.1 | 0.4 | GO:0090554 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.2 | GO:0033765 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.5 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.6 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.2 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 1.3 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.0 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.4 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 0.1 | GO:0015026 | coreceptor activity(GO:0015026) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.3 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.5 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.2 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.3 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 1.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.8 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |