Project
DANIO-CODE
Navigation
Downloads
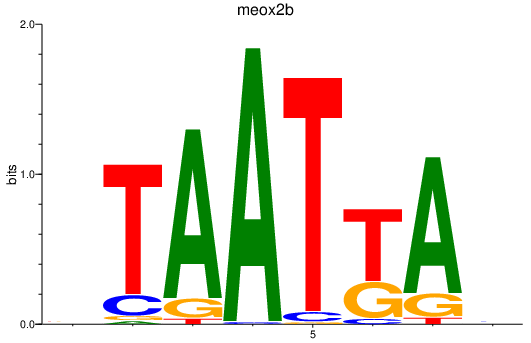
Results for meox2b
Z-value: 0.71
Transcription factors associated with meox2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
meox2b
|
ENSDARG00000061818 | mesenchyme homeobox 2b |
Activity profile of meox2b motif
Sorted Z-values of meox2b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of meox2b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_56157608 | 2.03 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr5_+_68410884 | 1.54 |
ENSDART00000153691
|
CU928129.1
|
ENSDARG00000097815 |
| chr10_+_39141022 | 1.51 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr14_+_46419051 | 1.33 |
ENSDART00000112377
|
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr3_+_56970554 | 1.30 |
ENSDART00000162930
|
bahcc1a
|
BAH domain and coiled-coil containing 1a |
| chr7_-_25623974 | 1.22 |
ENSDART00000173602
|
cd99l2
|
CD99 molecule-like 2 |
| chr11_-_29910947 | 1.20 |
ENSDART00000156121
|
scml2
|
sex comb on midleg-like 2 (Drosophila) |
| chr16_+_29060022 | 1.16 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr16_+_23516127 | 1.16 |
ENSDART00000004679
|
icn
|
ictacalcin |
| chr25_+_14411153 | 1.15 |
ENSDART00000015681
|
dbx1b
|
developing brain homeobox 1b |
| chr20_-_9107294 | 1.15 |
ENSDART00000140792
|
OMA1
|
OMA1 zinc metallopeptidase |
| chr23_-_12310778 | 1.13 |
ENSDART00000131256
|
phactr3a
|
phosphatase and actin regulator 3a |
| chr19_-_43037791 | 1.12 |
ENSDART00000004392
|
fkbp9
|
FK506 binding protein 9 |
| chr2_+_33343287 | 1.12 |
ENSDART00000056655
|
klf17
|
Kruppel-like factor 17 |
| chr2_+_32036450 | 1.10 |
ENSDART00000140776
|
CR391940.1
|
ENSDARG00000091946 |
| chr7_-_52848084 | 1.05 |
ENSDART00000172179
ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr16_+_13928844 | 1.01 |
ENSDART00000090191
|
flcn
|
folliculin |
| chr11_+_34788205 | 1.01 |
ENSDART00000124800
|
fam212aa
|
family with sequence similarity 212, member Aa |
| chr24_-_21778717 | 0.99 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr25_+_31457309 | 0.98 |
ENSDART00000022325
|
parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr22_-_15567180 | 0.96 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr4_-_2615160 | 0.94 |
ENSDART00000140760
|
e2f7
|
E2F transcription factor 7 |
| chr24_+_5205878 | 0.91 |
ENSDART00000106488
ENSDART00000005901 |
plod2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr11_-_2107054 | 0.89 |
ENSDART00000173031
|
hoxc6b
|
homeobox C6b |
| chr24_+_17125429 | 0.87 |
ENSDART00000017605
|
spag6
|
sperm associated antigen 6 |
| chr6_+_3873114 | 0.87 |
ENSDART00000159952
|
sema5bb
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Bb |
| chr3_+_17387551 | 0.86 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr23_+_19887319 | 0.86 |
ENSDART00000139192
|
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr9_+_25966093 | 0.86 |
ENSDART00000146011
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr13_+_35213326 | 0.85 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr21_-_22510751 | 0.84 |
ENSDART00000169870
|
myo5b
|
myosin VB |
| chr24_-_6048914 | 0.84 |
ENSDART00000146830
ENSDART00000021981 |
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr19_-_7576069 | 0.83 |
ENSDART00000148836
|
rfx5
|
regulatory factor X, 5 |
| chr1_-_57300743 | 0.83 |
ENSDART00000081122
|
COLGALT1 (1 of many)
|
collagen beta(1-O)galactosyltransferase 1 |
| chr4_+_3970874 | 0.83 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr19_+_7234029 | 0.82 |
ENSDART00000080348
|
brd2a
|
bromodomain containing 2a |
| chr25_-_30845998 | 0.81 |
ENSDART00000027661
|
myod1
|
myogenic differentiation 1 |
| chr1_-_40536800 | 0.81 |
ENSDART00000134739
|
rgs12b
|
regulator of G protein signaling 12b |
| chr13_+_27102377 | 0.80 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr8_+_2428689 | 0.80 |
ENSDART00000081325
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr13_+_27102308 | 0.80 |
ENSDART00000145901
|
rin2
|
Ras and Rab interactor 2 |
| chr6_-_23193752 | 0.79 |
ENSDART00000159749
|
usp43a
|
ubiquitin specific peptidase 43a |
| chr8_+_21321765 | 0.79 |
ENSDART00000131691
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr2_+_25468904 | 0.78 |
ENSDART00000056801
|
msl2a
|
male-specific lethal 2 homolog a (Drosophila) |
| chr1_+_32935645 | 0.77 |
ENSDART00000170832
|
arl13b
|
ADP-ribosylation factor-like 13b |
| chr6_+_47425082 | 0.77 |
ENSDART00000171087
|
si:ch211-286o17.1
|
si:ch211-286o17.1 |
| chr4_-_22751641 | 0.77 |
ENSDART00000066903
ENSDART00000130072 ENSDART00000123369 |
kmt2e
|
lysine (K)-specific methyltransferase 2E |
| chr24_-_39722595 | 0.76 |
ENSDART00000066506
|
cox6b1
|
cytochrome c oxidase subunit VIb polypeptide 1 |
| chr9_-_41608298 | 0.76 |
|
|
|
| chr18_-_782643 | 0.76 |
ENSDART00000161084
|
si:dkey-205h23.1
|
si:dkey-205h23.1 |
| chr10_+_39140943 | 0.76 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr2_+_49358871 | 0.75 |
ENSDART00000179089
|
sema6ba
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Ba |
| chr6_+_16341787 | 0.75 |
ENSDART00000114667
|
zgc:161969
|
zgc:161969 |
| chr22_-_22846670 | 0.75 |
ENSDART00000176355
|
CABZ01039424.2
|
ENSDARG00000108442 |
| chr4_-_59767735 | 0.75 |
ENSDART00000159114
|
znf1129
|
zinc finger protein 1129 |
| chr9_-_48098629 | 0.75 |
|
|
|
| chr21_+_26684617 | 0.75 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr1_+_13779755 | 0.74 |
|
|
|
| chr5_-_40707316 | 0.74 |
ENSDART00000161932
|
npr3
|
natriuretic peptide receptor 3 |
| chr23_-_23474703 | 0.74 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr16_-_17439735 | 0.73 |
ENSDART00000144392
|
zyx
|
zyxin |
| chr21_-_8420626 | 0.73 |
ENSDART00000084378
|
crb2a
|
crumbs family member 2a |
| chr22_-_26333957 | 0.73 |
ENSDART00000130493
|
capn2b
|
calpain 2, (m/II) large subunit b |
| chr17_-_14868764 | 0.73 |
ENSDART00000115064
|
nid2a
|
nidogen 2a (osteonidogen) |
| chr2_-_34010299 | 0.73 |
ENSDART00000140910
|
ptch2
|
patched 2 |
| chr14_+_20045365 | 0.72 |
ENSDART00000167637
|
aff2
|
AF4/FMR2 family, member 2 |
| chr11_-_44724371 | 0.72 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr8_-_22945616 | 0.71 |
|
|
|
| chr7_-_33921565 | 0.71 |
ENSDART00000125131
|
smad6a
|
SMAD family member 6a |
| chr12_-_5693715 | 0.71 |
ENSDART00000105887
|
dlx4b
|
distal-less homeobox 4b |
| chr19_+_22478256 | 0.71 |
ENSDART00000100181
|
sall3b
|
spalt-like transcription factor 3b |
| chr16_+_46145286 | 0.70 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr16_+_11138924 | 0.70 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr20_-_35343057 | 0.70 |
ENSDART00000113294
|
fzd3a
|
frizzled class receptor 3a |
| chr10_-_15896595 | 0.70 |
ENSDART00000092343
|
tjp2a
|
tight junction protein 2a (zona occludens 2) |
| chr21_-_25567978 | 0.69 |
ENSDART00000133134
|
efemp2b
|
EGF containing fibulin-like extracellular matrix protein 2b |
| chr13_+_27654900 | 0.68 |
ENSDART00000160550
|
CR356236.1
|
ENSDARG00000102040 |
| chr9_-_18903879 | 0.68 |
ENSDART00000138785
|
si:dkey-239h2.3
|
si:dkey-239h2.3 |
| chr23_+_6043862 | 0.68 |
|
|
|
| chr11_+_37639045 | 0.68 |
ENSDART00000111157
|
si:ch211-112f3.4
|
si:ch211-112f3.4 |
| chr11_+_14142126 | 0.68 |
ENSDART00000102520
|
palm1a
|
paralemmin 1a |
| chr13_+_2774422 | 0.67 |
ENSDART00000162362
|
wu:fj16a03
|
wu:fj16a03 |
| chr8_-_17480730 | 0.67 |
ENSDART00000100667
|
skia
|
v-ski avian sarcoma viral oncogene homolog a |
| chr1_-_23765358 | 0.67 |
|
|
|
| chr7_+_34018862 | 0.67 |
ENSDART00000123498
|
fibinb
|
fin bud initiation factor b |
| chr3_-_23513177 | 0.67 |
ENSDART00000078425
|
eve1
|
even-skipped-like1 |
| chr24_+_38371710 | 0.66 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr16_+_24046472 | 0.66 |
|
|
|
| chr20_-_36984259 | 0.66 |
ENSDART00000076313
|
txlnba
|
taxilin beta a |
| chr10_-_17630376 | 0.66 |
ENSDART00000113101
|
smarcad1b
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 b |
| chr24_+_16998329 | 0.65 |
ENSDART00000177272
|
mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr7_-_32888309 | 0.65 |
ENSDART00000173461
|
BX571811.2
|
ENSDARG00000105655 |
| chr4_-_73485204 | 0.65 |
ENSDART00000122898
|
phf21b
|
PHD finger protein 21B |
| chr20_-_21031504 | 0.64 |
ENSDART00000152726
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr15_-_18159904 | 0.64 |
ENSDART00000170874
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr2_+_27344633 | 0.64 |
ENSDART00000178275
|
cdh7
|
cadherin 7, type 2 |
| chr3_+_18249107 | 0.63 |
ENSDART00000141100
ENSDART00000138107 |
rps2
|
ribosomal protein S2 |
| chr15_+_29343644 | 0.63 |
ENSDART00000170537
ENSDART00000128973 |
rap1gap2a
|
RAP1 GTPase activating protein 2a |
| chr12_+_3043444 | 0.63 |
ENSDART00000149427
|
sgca
|
sarcoglycan, alpha |
| chr12_+_20230575 | 0.63 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr12_-_4497094 | 0.63 |
ENSDART00000163651
|
trpm4b.2
|
transient receptor potential cation channel, subfamily M, member 4b, transient receptor potential cation channel, subfamily M, member 4b, tandem duplicate 2 |
| chr16_-_7525980 | 0.63 |
ENSDART00000017445
|
prdm1a
|
PR domain containing 1a, with ZNF domain |
| chr10_+_42530040 | 0.62 |
ENSDART00000025691
|
dbnla
|
drebrin-like a |
| chr20_-_3970778 | 0.62 |
ENSDART00000178724
ENSDART00000178565 |
TRIM67
|
tripartite motif containing 67 |
| chr15_-_28663537 | 0.62 |
ENSDART00000156800
|
si:ch211-225b7.5
|
si:ch211-225b7.5 |
| chr5_+_26925238 | 0.61 |
ENSDART00000051491
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr16_+_11138879 | 0.61 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr17_+_23278879 | 0.61 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr8_+_49789789 | 0.61 |
ENSDART00000083790
|
ntrk2a
|
neurotrophic tyrosine kinase, receptor, type 2a |
| chr9_-_20562293 | 0.61 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr15_-_1858350 | 0.61 |
ENSDART00000082026
|
mmp28
|
matrix metallopeptidase 28 |
| chr17_+_15425559 | 0.60 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr19_+_12995955 | 0.60 |
ENSDART00000132892
|
cthrc1a
|
collagen triple helix repeat containing 1a |
| chr16_-_45269179 | 0.60 |
ENSDART00000162095
|
si:dkey-33i11.4
|
si:dkey-33i11.4 |
| chr19_-_31815128 | 0.60 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr2_-_23348612 | 0.59 |
ENSDART00000110373
|
znf414
|
zinc finger protein 414 |
| chr18_+_5899355 | 0.59 |
|
|
|
| chr2_+_57903155 | 0.59 |
|
|
|
| chr25_+_30699938 | 0.59 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr7_-_72278552 | 0.59 |
ENSDART00000168532
|
HECTD4
|
HECT domain E3 ubiquitin protein ligase 4 |
| chr21_+_17731439 | 0.59 |
ENSDART00000124173
|
rxraa
|
retinoid X receptor, alpha a |
| chr18_-_3056732 | 0.59 |
ENSDART00000162657
|
rps3
|
ribosomal protein S3 |
| chr4_-_15442828 | 0.59 |
ENSDART00000157414
|
plxna4
|
plexin A4 |
| chr4_+_18974767 | 0.59 |
ENSDART00000066973
|
impdh1b
|
IMP (inosine 5'-monophosphate) dehydrogenase 1b |
| chr8_+_30443651 | 0.58 |
ENSDART00000062303
|
foxd5
|
forkhead box D5 |
| chr4_-_51772774 | 0.58 |
|
|
|
| chr23_-_22186674 | 0.57 |
ENSDART00000142474
|
phc2a
|
polyhomeotic homolog 2a (Drosophila) |
| chr17_+_24668907 | 0.57 |
ENSDART00000034263
ENSDART00000135794 |
sepn1
|
selenoprotein N, 1 |
| chr15_-_14616083 | 0.57 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr17_-_29102320 | 0.57 |
ENSDART00000104204
|
foxg1a
|
forkhead box G1a |
| chr22_-_15930756 | 0.56 |
ENSDART00000080047
|
eps15l1a
|
epidermal growth factor receptor pathway substrate 15-like 1a |
| chr2_+_44692913 | 0.56 |
ENSDART00000155362
|
ENSDARG00000079124
|
ENSDARG00000079124 |
| chr6_-_54816183 | 0.56 |
ENSDART00000148462
|
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr22_-_24341408 | 0.56 |
|
|
|
| chr13_-_31165867 | 0.56 |
ENSDART00000030946
|
prdm8
|
PR domain containing 8 |
| chr6_-_40314512 | 0.55 |
ENSDART00000033844
|
col7a1
|
collagen, type VII, alpha 1 |
| chr16_-_28943421 | 0.55 |
ENSDART00000149501
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr5_-_64149931 | 0.54 |
ENSDART00000144816
|
lix1
|
limb and CNS expressed 1 |
| chr8_+_2260519 | 0.54 |
ENSDART00000136743
|
ikbkb
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta |
| chr8_-_17580655 | 0.54 |
|
|
|
| chr3_-_59884282 | 0.54 |
ENSDART00000156597
|
si:ch73-364h19.1
|
si:ch73-364h19.1 |
| chr6_-_55881470 | 0.54 |
ENSDART00000160991
|
cyp24a1
|
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chr8_-_23591082 | 0.54 |
ENSDART00000025024
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr22_+_9889203 | 0.53 |
ENSDART00000177953
|
blf
|
bloody fingers |
| chr3_+_28822408 | 0.53 |
ENSDART00000133528
|
lgals2a
|
lectin, galactoside-binding, soluble, 2a |
| chr11_-_17620732 | 0.52 |
ENSDART00000154627
|
eogt
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr3_-_60929921 | 0.52 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr18_+_618393 | 0.52 |
ENSDART00000159464
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr22_+_2921305 | 0.52 |
ENSDART00000143258
|
cep19
|
centrosomal protein 19 |
| chr14_-_4166292 | 0.52 |
ENSDART00000127318
|
frmpd1b
|
FERM and PDZ domain containing 1b |
| chr11_+_29753417 | 0.52 |
|
|
|
| chr1_+_16681778 | 0.52 |
|
|
|
| chr6_+_54231519 | 0.52 |
ENSDART00000149542
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr20_-_29517770 | 0.51 |
ENSDART00000147464
|
ryr3
|
ryanodine receptor 3 |
| chr3_-_60866373 | 0.51 |
ENSDART00000112043
|
cacng4b
|
calcium channel, voltage-dependent, gamma subunit 4b |
| chr15_+_42329269 | 0.51 |
ENSDART00000099234
ENSDART00000152731 |
scaf4b
|
SR-related CTD-associated factor 4b |
| chr7_+_30516734 | 0.51 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr8_+_46378250 | 0.51 |
ENSDART00000129661
ENSDART00000084081 |
ogg1
|
8-oxoguanine DNA glycosylase |
| chr15_-_734338 | 0.51 |
ENSDART00000155878
|
si:dkey-7i4.23
|
si:dkey-7i4.23 |
| chr23_+_11350727 | 0.51 |
|
|
|
| chr22_-_36561247 | 0.50 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr14_-_45883839 | 0.50 |
|
|
|
| chr9_-_35205969 | 0.50 |
|
|
|
| chr6_+_50393779 | 0.50 |
ENSDART00000055502
|
ergic3
|
ERGIC and golgi 3 |
| chr6_-_43273456 | 0.49 |
|
|
|
| chr22_-_864745 | 0.49 |
ENSDART00000035514
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr24_+_7832020 | 0.49 |
ENSDART00000019705
|
bmp6
|
bone morphogenetic protein 6 |
| chr2_+_26632673 | 0.49 |
ENSDART00000017668
|
ptbp1a
|
polypyrimidine tract binding protein 1a |
| chr24_+_35500964 | 0.49 |
ENSDART00000058571
|
snai2
|
snail family zinc finger 2 |
| chr15_+_880573 | 0.49 |
ENSDART00000115077
|
zgc:174573
|
zgc:174573 |
| chr15_-_40391882 | 0.49 |
|
|
|
| chr2_-_32370176 | 0.49 |
ENSDART00000077151
|
asap1a
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1a |
| chr25_-_5835981 | 0.48 |
ENSDART00000155751
|
nuak1b
|
NUAK family, SNF1-like kinase, 1b |
| chr4_+_14728248 | 0.48 |
ENSDART00000124189
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr4_+_9668755 | 0.48 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr19_+_19939410 | 0.48 |
|
|
|
| chr20_+_55096127 | 0.48 |
|
|
|
| chr13_+_33151628 | 0.48 |
ENSDART00000135200
|
ccdc28b
|
coiled-coil domain containing 28B |
| chr2_-_23516930 | 0.48 |
ENSDART00000165355
|
prrx1a
|
paired related homeobox 1a |
| chr17_-_25630666 | 0.48 |
ENSDART00000149060
|
ppp1cb
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr7_+_54410044 | 0.47 |
|
|
|
| chr21_+_28408329 | 0.47 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr21_+_25199691 | 0.47 |
ENSDART00000168140
ENSDART00000112783 |
tmem45b
|
transmembrane protein 45B |
| chr9_-_56292487 | 0.47 |
ENSDART00000151720
|
si:ch211-39i22.1
|
si:ch211-39i22.1 |
| chr22_+_16509286 | 0.47 |
ENSDART00000083063
|
tal1
|
T-cell acute lymphocytic leukemia 1 |
| chr11_-_6442836 | 0.47 |
ENSDART00000004483
|
zgc:162969
|
zgc:162969 |
| chr1_-_40587797 | 0.47 |
ENSDART00000170895
|
rgs12b
|
regulator of G protein signaling 12b |
| chr9_+_25966225 | 0.47 |
ENSDART00000146011
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr16_+_43249142 | 0.47 |
ENSDART00000154493
|
adam22
|
ADAM metallopeptidase domain 22 |
| chr22_+_223797 | 0.47 |
|
|
|
| chr12_+_24221087 | 0.47 |
ENSDART00000088178
|
nrxn1a
|
neurexin 1a |
| chr21_+_10663517 | 0.46 |
ENSDART00000074833
|
rx3
|
retinal homeobox gene 3 |
| chr7_-_72500748 | 0.46 |
ENSDART00000160523
|
CU929444.1
|
ENSDARG00000099109 |
| chr22_-_36783601 | 0.46 |
|
|
|
| chr24_-_14447773 | 0.46 |
|
|
|
| chr12_+_1572834 | 0.45 |
|
|
|
| chr2_-_10919978 | 0.45 |
ENSDART00000005944
|
rpl5a
|
ribosomal protein L5a |
| chr9_-_12453581 | 0.45 |
ENSDART00000088199
|
zgc:162707
|
zgc:162707 |
| chr21_-_17259886 | 0.45 |
ENSDART00000114877
|
gfi1b
|
growth factor independent 1B transcription repressor |
| chr3_-_21217799 | 0.45 |
ENSDART00000114906
|
fam171a2a
|
family with sequence similarity 171, member A2a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.3 | 0.9 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.3 | 0.9 | GO:0001659 | temperature homeostasis(GO:0001659) sleep(GO:0030431) |
| 0.3 | 0.8 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.3 | 1.1 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.3 | 1.0 | GO:0086014 | atrial cardiac muscle cell action potential(GO:0086014) cell-cell signaling involved in cardiac conduction(GO:0086019) atrial cardiac muscle cell to AV node cell signaling(GO:0086026) cell communication involved in cardiac conduction(GO:0086065) atrial cardiac muscle cell to AV node cell communication(GO:0086066) |
| 0.3 | 0.8 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.2 | 1.3 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.2 | 1.0 | GO:0003379 | establishment of cell polarity involved in gastrulation cell migration(GO:0003379) |
| 0.2 | 0.6 | GO:0002706 | regulation of lymphocyte mediated immunity(GO:0002706) |
| 0.2 | 0.6 | GO:0051155 | positive regulation of striated muscle cell differentiation(GO:0051155) |
| 0.2 | 0.6 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.2 | 0.6 | GO:1902024 | serine transport(GO:0032329) L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.2 | 0.8 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.2 | 0.7 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.2 | 0.5 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.2 | 0.8 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.2 | 0.8 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.2 | 0.6 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.2 | 0.5 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.1 | 0.4 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) microglia development(GO:0014005) regulation of cellular amine metabolic process(GO:0033238) |
| 0.1 | 0.7 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.1 | 2.4 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.1 | 0.7 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 1.4 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 0.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.5 | GO:0033280 | response to vitamin D(GO:0033280) vitamin D metabolic process(GO:0042359) |
| 0.1 | 0.5 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 0.4 | GO:0052803 | imidazole-containing compound metabolic process(GO:0052803) imidazole-containing compound catabolic process(GO:0052805) |
| 0.1 | 0.5 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.9 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.1 | 0.3 | GO:0034433 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.1 | 0.7 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.1 | 0.4 | GO:0010662 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.1 | 0.4 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.1 | 0.4 | GO:0046443 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.1 | 0.3 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.1 | 0.8 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 0.8 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.9 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.1 | 0.4 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.4 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.1 | 2.7 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 0.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.8 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.1 | 0.6 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 0.6 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 1.0 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 0.5 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.1 | 0.4 | GO:0007622 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.1 | 0.7 | GO:0070307 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 0.4 | GO:0045056 | transcytosis(GO:0045056) |
| 0.1 | 0.2 | GO:0046888 | negative regulation of hormone secretion(GO:0046888) |
| 0.1 | 0.4 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 0.3 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.7 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 0.7 | GO:0046037 | GMP biosynthetic process(GO:0006177) GTP biosynthetic process(GO:0006183) GMP metabolic process(GO:0046037) |
| 0.1 | 0.4 | GO:0035637 | multicellular organismal signaling(GO:0035637) |
| 0.1 | 0.6 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 0.3 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 1.0 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 0.2 | GO:0051414 | response to cortisol(GO:0051414) cellular response to cortisol stimulus(GO:0071387) |
| 0.1 | 0.2 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.1 | 0.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.2 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 1.0 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 0.3 | GO:1900028 | negative regulation of Rac protein signal transduction(GO:0035021) wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.6 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 0.7 | GO:0030656 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.1 | 0.2 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 0.1 | GO:0003207 | cardiac chamber formation(GO:0003207) cardiac ventricle formation(GO:0003211) |
| 0.1 | 0.3 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 0.9 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 2.1 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.1 | 0.2 | GO:1990481 | snRNA pseudouridine synthesis(GO:0031120) mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.3 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.0 | 0.5 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.3 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.7 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.2 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.2 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 0.3 | GO:0036268 | swimming(GO:0036268) |
| 0.0 | 0.2 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.5 | GO:0003313 | heart rudiment development(GO:0003313) |
| 0.0 | 0.2 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.1 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.7 | GO:0055017 | cardiac muscle tissue growth(GO:0055017) |
| 0.0 | 0.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.6 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.4 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.2 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.5 | GO:0044247 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.2 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.1 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.4 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.4 | GO:0030241 | skeletal muscle thin filament assembly(GO:0030240) skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.8 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.4 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.2 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.0 | 0.4 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 0.8 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.7 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.6 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.1 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.2 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.4 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.6 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 1.0 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.6 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.9 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.0 | 1.4 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.3 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.1 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.2 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.0 | 0.2 | GO:0048696 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.2 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.3 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.1 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.1 | GO:1902868 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.0 | 0.0 | GO:0003342 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) |
| 0.0 | 0.4 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.2 | GO:0006956 | complement activation(GO:0006956) |
| 0.0 | 0.2 | GO:0035437 | maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 1.1 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.0 | 0.5 | GO:0039020 | pronephric nephron tubule development(GO:0039020) |
| 0.0 | 0.6 | GO:0032231 | regulation of actin filament bundle assembly(GO:0032231) |
| 0.0 | 0.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.7 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.0 | 0.6 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.1 | GO:0071422 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.0 | 0.3 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 0.6 | GO:0097581 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.0 | 0.7 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.5 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.3 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.2 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.8 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.2 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.3 | GO:0035675 | neuromast hair cell development(GO:0035675) |
| 0.0 | 0.1 | GO:0036230 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.0 | 0.1 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.0 | 0.4 | GO:0015696 | ammonium transport(GO:0015696) |
| 0.0 | 0.5 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.0 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.0 | 0.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.4 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.3 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.3 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.3 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 1.0 | GO:0001708 | cell fate specification(GO:0001708) |
| 0.0 | 0.1 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.0 | GO:0035909 | aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.0 | 0.1 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.7 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.3 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.6 | GO:0018210 | peptidyl-threonine modification(GO:0018210) |
| 0.0 | 0.1 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) positive regulation of organelle assembly(GO:1902117) |
| 0.0 | 0.3 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.1 | GO:0060173 | limb development(GO:0060173) |
| 0.0 | 0.3 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.4 | GO:1903844 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.0 | GO:0032475 | otolith formation(GO:0032475) |
| 0.0 | 0.2 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.4 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.3 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.4 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 0.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.8 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 1.0 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.6 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.4 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.1 | 0.6 | GO:0031838 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.6 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.5 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.1 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.8 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.7 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.4 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.1 | 0.8 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 1.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.4 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.2 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 1.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.4 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.7 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 1.2 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 2.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.8 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 2.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.2 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.8 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.4 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.7 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 1.0 | GO:0005604 | basement membrane(GO:0005604) extracellular matrix component(GO:0044420) |
| 0.0 | 0.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.2 | GO:0030677 | nucleoid(GO:0009295) ribonuclease P complex(GO:0030677) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0031674 | I band(GO:0031674) |
| 0.0 | 0.0 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.3 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 1.5 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.4 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.3 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 0.0 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.0 | 0.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0044304 | main axon(GO:0044304) |
| 0.0 | 0.4 | GO:0014069 | postsynaptic density(GO:0014069) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.2 | 0.7 | GO:0008158 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.2 | 0.5 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.2 | 0.5 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.2 | 0.6 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.7 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 0.4 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.4 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.1 | 0.4 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.5 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.8 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 0.5 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 0.4 | GO:0016841 | folic acid binding(GO:0005542) ammonia-lyase activity(GO:0016841) |
| 0.1 | 0.7 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.4 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.1 | 1.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.3 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.1 | 0.4 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.1 | 0.5 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.5 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.6 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.4 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.4 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.1 | 0.3 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.1 | 0.6 | GO:0015149 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.1 | 0.4 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 0.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.3 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.6 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.5 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 0.8 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 0.4 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.3 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.3 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.1 | 0.2 | GO:0019809 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.1 | 0.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 1.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.2 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.0 | 0.7 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.3 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.9 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.6 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.4 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.3 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 1.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.2 | GO:0019869 | chloride channel regulator activity(GO:0017081) chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 1.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 1.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 1.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.4 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.1 | GO:0035381 | purinergic nucleotide receptor activity(GO:0001614) extracellular ATP-gated cation channel activity(GO:0004931) nucleotide receptor activity(GO:0016502) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 1.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.3 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.8 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.7 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 5.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0015141 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.3 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.4 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.9 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 1.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.6 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.0 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.1 | GO:0004691 | AMP-activated protein kinase activity(GO:0004679) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.5 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 2.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.2 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0008009 | chemokine activity(GO:0008009) chemokine receptor binding(GO:0042379) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.0 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 2.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 0.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 0.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.3 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.4 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.1 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 2.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.4 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.3 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 0.8 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 0.8 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 0.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 0.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 0.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 0.6 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.3 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.0 | 1.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.8 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 2.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.4 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.5 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.3 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.4 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |