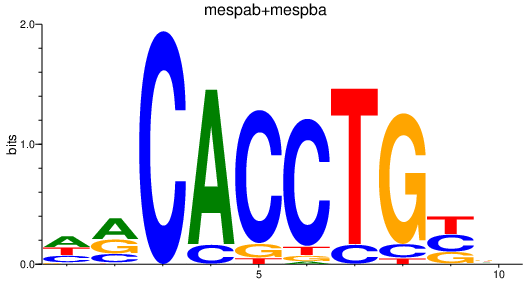
Project
DANIO-CODE
Navigation
Downloads
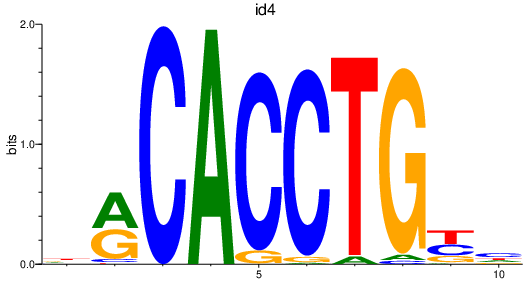
Results for mespab+mespba_id4
Z-value: 3.60
Transcription factors associated with mespab+mespba_id4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mespba
|
ENSDARG00000030347 | mesoderm posterior ba |
|
mespab
|
ENSDARG00000068761 | mesoderm posterior ab |
|
id4
|
ENSDARG00000045131 | inhibitor of DNA binding 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| id4 | dr10_dc_chr16_-_10089440_10089472 | -0.81 | 1.3e-04 | Click! |
| mespba | dr10_dc_chr7_+_15059782_15059806 | 0.78 | 3.9e-04 | Click! |
| mespab | dr10_dc_chr25_-_10919875_10919883 | -0.59 | 1.5e-02 | Click! |
Activity profile of mespab+mespba_id4 motif
Sorted Z-values of mespab+mespba_id4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of mespab+mespba_id4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_58448953 | 25.24 |
ENSDART00000024185
|
zgc:56231
|
zgc:56231 |
| chr21_-_30256477 | 17.18 |
ENSDART00000137193
|
slbp2
|
stem-loop binding protein 2 |
| chr21_-_34227158 | 14.67 |
ENSDART00000169218
ENSDART00000064320 ENSDART00000172381 |
alg13
|
ALG13, UDP-N-acetylglucosaminyltransferase subunit |
| chr11_+_30005768 | 14.59 |
ENSDART00000167618
|
CR790368.1
|
ENSDARG00000100936 |
| chr2_+_28199458 | 14.47 |
ENSDART00000150330
|
buc
|
bucky ball |
| chr4_-_20456825 | 13.55 |
ENSDART00000003621
ENSDART00000132356 |
sinup
|
siaz-interacting nuclear protein |
| chr7_+_58449163 | 13.28 |
ENSDART00000167166
|
zgc:56231
|
zgc:56231 |
| chr12_+_13053552 | 13.15 |
ENSDART00000124799
|
si:ch211-103b1.2
|
si:ch211-103b1.2 |
| chr13_-_21541531 | 13.00 |
ENSDART00000067537
|
elovl6l
|
ELOVL family member 6, elongation of long chain fatty acids like |
| chr23_+_540624 | 12.29 |
ENSDART00000034707
|
lsm14b
|
LSM family member 14B |
| chr10_-_25246786 | 11.53 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr15_+_38397772 | 10.81 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr7_+_58449127 | 10.77 |
ENSDART00000167166
|
zgc:56231
|
zgc:56231 |
| chr12_-_10474942 | 10.63 |
ENSDART00000106163
ENSDART00000124562 |
zgc:152977
|
zgc:152977 |
| chr23_+_2786407 | 10.59 |
ENSDART00000066086
|
zgc:114123
|
zgc:114123 |
| chr1_-_33717934 | 10.19 |
ENSDART00000083736
|
lmo7b
|
LIM domain 7b |
| chr16_-_42116471 | 9.97 |
ENSDART00000058620
|
zp3d.1
|
zona pellucida glycoprotein 3d tandem duplicate 1 |
| chr6_+_135002 | 9.84 |
ENSDART00000097468
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr22_+_17235696 | 9.72 |
ENSDART00000134798
|
tdrd5
|
tudor domain containing 5 |
| chr15_-_43283562 | 9.56 |
ENSDART00000110352
|
ap1s3a
|
adaptor-related protein complex 1, sigma 3 subunit, a |
| chr3_+_42217236 | 9.25 |
ENSDART00000168228
|
tmem184a
|
transmembrane protein 184a |
| chr7_-_47978449 | 9.25 |
ENSDART00000127007
ENSDART00000024062 |
cpeb1b
|
cytoplasmic polyadenylation element binding protein 1b |
| chr19_+_15536640 | 9.20 |
ENSDART00000098970
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr7_-_55346967 | 9.19 |
ENSDART00000135304
|
pabpn1l
|
poly(A) binding protein, nuclear 1-like (cytoplasmic) |
| chr24_-_33894062 | 9.17 |
ENSDART00000079210
|
cdk5
|
cyclin-dependent kinase 5 |
| chr2_-_50491234 | 9.05 |
ENSDART00000165678
|
mcm6l
|
MCM6 minichromosome maintenance deficient 6, like |
| chr7_+_67475765 | 8.98 |
ENSDART00000160086
|
zgc:162592
|
zgc:162592 |
| chr12_+_30389706 | 8.97 |
|
|
|
| chr2_-_47766563 | 8.83 |
ENSDART00000038228
|
ap1s3b
|
adaptor-related protein complex 1, sigma 3 subunit, b |
| chr10_+_16914003 | 8.72 |
ENSDART00000177906
|
UNC13B
|
unc-13 homolog B |
| chr19_+_14592632 | 8.65 |
ENSDART00000161088
ENSDART00000161965 |
zdhhc18b
|
zinc finger, DHHC-type containing 18b |
| chr1_-_18118467 | 8.54 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr8_+_45326435 | 8.38 |
ENSDART00000134161
|
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr5_-_31738565 | 8.25 |
ENSDART00000017956
|
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr12_+_13704712 | 7.80 |
ENSDART00000152257
|
ppp1r16a
|
protein phosphatase 1, regulatory subunit 16A |
| chr7_+_58448909 | 7.78 |
ENSDART00000024185
|
zgc:56231
|
zgc:56231 |
| chr14_-_32937496 | 7.72 |
ENSDART00000048130
|
stard14
|
START domain containing 14 |
| chr18_-_25784873 | 7.58 |
ENSDART00000103046
|
zgc:162879
|
zgc:162879 |
| chr6_+_40525779 | 7.36 |
ENSDART00000033819
|
prkcda
|
protein kinase C, delta a |
| chr21_-_804453 | 7.30 |
|
|
|
| chr12_+_22551105 | 7.23 |
ENSDART00000123808
ENSDART00000159864 |
cdca9
|
cell division cycle associated 9 |
| chr17_-_36913213 | 7.10 |
ENSDART00000154981
|
senp6b
|
SUMO1/sentrin specific peptidase 6b |
| chr5_+_57254393 | 6.97 |
ENSDART00000050949
|
btg4
|
B-cell translocation gene 4 |
| chr7_+_56787468 | 6.92 |
ENSDART00000024076
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr5_-_14993912 | 6.91 |
ENSDART00000085943
|
taok3a
|
TAO kinase 3a |
| chr16_-_47446494 | 6.80 |
ENSDART00000032188
|
si:dkey-256h2.1
|
si:dkey-256h2.1 |
| chr2_+_6341404 | 6.80 |
ENSDART00000076700
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr22_+_25113293 | 6.70 |
ENSDART00000171851
|
si:ch211-226h8.4
|
si:ch211-226h8.4 |
| chr10_+_29247498 | 6.60 |
|
|
|
| chr5_-_23192997 | 6.45 |
ENSDART00000167629
|
rnf128a
|
ring finger protein 128a |
| chr25_-_9889107 | 6.42 |
ENSDART00000137407
|
AL929493.1
|
ENSDARG00000093575 |
| chr5_+_44246311 | 6.40 |
ENSDART00000145299
ENSDART00000136521 |
kank1a
|
KN motif and ankyrin repeat domains 1a |
| chr22_+_22413803 | 6.37 |
ENSDART00000147825
|
kif14
|
kinesin family member 14 |
| chr3_-_39920399 | 6.29 |
|
|
|
| chr15_+_29092022 | 6.28 |
ENSDART00000141164
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr12_+_20569659 | 6.19 |
ENSDART00000141804
|
st6galnac1.2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1, tandem duplicate 2 |
| chr9_-_34459799 | 6.17 |
ENSDART00000059955
|
ildr1b
|
immunoglobulin-like domain containing receptor 1b |
| chr14_-_21320696 | 6.13 |
ENSDART00000043162
|
reep2
|
receptor accessory protein 2 |
| chr23_+_6652454 | 6.00 |
ENSDART00000081763
|
rbm38
|
RNA binding motif protein 38 |
| chr2_-_54224744 | 5.99 |
|
|
|
| chr22_-_25592743 | 5.86 |
ENSDART00000110638
ENSDART00000172092 |
si:ch211-12h2.8
|
si:ch211-12h2.8 |
| chr17_-_25313024 | 5.79 |
ENSDART00000082324
|
zpcx
|
zona pellucida protein C |
| chr22_-_24964889 | 5.64 |
ENSDART00000102751
|
si:dkey-179j5.5
|
si:dkey-179j5.5 |
| chr14_-_32937536 | 5.64 |
ENSDART00000132850
|
stard14
|
START domain containing 14 |
| chr15_+_29092224 | 5.60 |
ENSDART00000131755
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr13_-_37530793 | 5.54 |
ENSDART00000141295
|
si:dkey-188i13.11
|
si:dkey-188i13.11 |
| chr2_-_44330306 | 5.53 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
| chr9_-_38213786 | 5.50 |
|
|
|
| chr20_-_46079578 | 5.34 |
ENSDART00000153228
|
AL929237.3
|
ENSDARG00000096676 |
| chr8_+_13327234 | 5.34 |
ENSDART00000159760
|
slc5a5
|
solute carrier family 5 (sodium/iodide cotransporter), member 5 |
| chr14_+_44703094 | 5.28 |
|
|
|
| chr5_-_64983648 | 5.27 |
ENSDART00000147707
|
camsap1b
|
calmodulin regulated spectrin-associated protein 1b |
| chr19_-_18664720 | 5.24 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| chr3_+_42217187 | 5.23 |
ENSDART00000168228
|
tmem184a
|
transmembrane protein 184a |
| chr4_-_20456437 | 5.20 |
ENSDART00000003621
ENSDART00000132356 |
sinup
|
siaz-interacting nuclear protein |
| chr19_-_7402373 | 5.18 |
ENSDART00000092375
|
oxr1b
|
oxidation resistance 1b |
| chr6_+_10098305 | 5.16 |
ENSDART00000151477
|
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr22_+_17803309 | 5.15 |
ENSDART00000136016
|
hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr2_+_1645259 | 5.14 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr20_-_13729816 | 5.13 |
ENSDART00000078893
|
ezrb
|
ezrin b |
| chr16_+_4350448 | 5.11 |
|
|
|
| chr17_+_699708 | 5.10 |
ENSDART00000165144
|
siva1
|
SIVA1, apoptosis-inducing factor |
| chr11_-_25615491 | 5.09 |
ENSDART00000145655
|
tmem51b
|
transmembrane protein 51b |
| chr23_+_39961676 | 5.06 |
ENSDART00000161881
|
ENSDARG00000104435
|
ENSDARG00000104435 |
| chr10_-_1933761 | 5.04 |
ENSDART00000101023
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr15_-_28963974 | 5.00 |
ENSDART00000155305
|
eml2
|
echinoderm microtubule associated protein like 2 |
| chr2_+_10858480 | 4.98 |
ENSDART00000091570
|
fam69aa
|
family with sequence similarity 69, member Aa |
| chr3_+_59972297 | 4.97 |
|
|
|
| chr5_-_23192934 | 4.96 |
ENSDART00000021462
|
rnf128a
|
ring finger protein 128a |
| chr5_-_64983687 | 4.95 |
ENSDART00000147707
|
camsap1b
|
calmodulin regulated spectrin-associated protein 1b |
| chr16_-_44179964 | 4.95 |
ENSDART00000058685
|
zfpm2a
|
zinc finger protein, FOG family member 2a |
| chr15_+_38397715 | 4.92 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr24_+_26223767 | 4.92 |
|
|
|
| chr24_+_14792755 | 4.86 |
ENSDART00000091735
|
dok6
|
docking protein 6 |
| chr23_-_31528548 | 4.82 |
|
|
|
| chr3_-_29779725 | 4.79 |
ENSDART00000151501
|
RUNDC1
|
RUN domain containing 1 |
| chr6_+_135255 | 4.76 |
ENSDART00000097468
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr13_-_9110703 | 4.76 |
ENSDART00000138833
|
grxcr1
|
glutaredoxin, cysteine rich 1 |
| chr9_+_38832959 | 4.74 |
ENSDART00000110651
|
slc12a8
|
solute carrier family 12, member 8 |
| chr12_+_46242558 | 4.67 |
ENSDART00000167510
|
hid1b
|
HID1 domain containing b |
| chr23_+_29017954 | 4.65 |
ENSDART00000140291
|
CR677513.1
|
ENSDARG00000093890 |
| chr3_-_29779598 | 4.62 |
ENSDART00000151501
|
RUNDC1
|
RUN domain containing 1 |
| chr12_+_47448318 | 4.61 |
ENSDART00000152857
|
fmn2b
|
formin 2b |
| chr2_+_25212281 | 4.61 |
ENSDART00000078838
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr1_+_18118735 | 4.60 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr14_+_31596468 | 4.58 |
ENSDART00000173259
|
BX005454.1
|
ENSDARG00000100646 |
| chr22_-_4787016 | 4.55 |
ENSDART00000140313
|
tnfaip8l1
|
tumor necrosis factor, alpha-induced protein 8-like 1 |
| chr19_+_43316957 | 4.48 |
ENSDART00000151298
|
arpp21
|
cAMP-regulated phosphoprotein, 21 |
| chr10_+_37229202 | 4.47 |
ENSDART00000136510
|
ksr1a
|
kinase suppressor of ras 1a |
| chr12_-_4264663 | 4.45 |
ENSDART00000152521
|
ca15b
|
carbonic anhydrase XVb |
| chr25_+_5845303 | 4.43 |
ENSDART00000163948
|
ENSDARG00000053246
|
ENSDARG00000053246 |
| chr20_-_46079529 | 4.42 |
ENSDART00000153228
|
AL929237.3
|
ENSDARG00000096676 |
| chr5_-_56953587 | 4.42 |
ENSDART00000167892
|
zgc:193711
|
zgc:193711 |
| chr14_-_47210912 | 4.37 |
ENSDART00000031640
ENSDART00000132966 |
zgc:113425
|
zgc:113425 |
| chr5_-_23211957 | 4.36 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr14_-_35932521 | 4.35 |
ENSDART00000158722
|
BX511223.1
|
ENSDARG00000101064 |
| chr1_+_41762057 | 4.26 |
ENSDART00000137609
|
ctnna2
|
catenin (cadherin-associated protein), alpha 2 |
| chr19_+_14592757 | 4.24 |
ENSDART00000161088
ENSDART00000161965 |
zdhhc18b
|
zinc finger, DHHC-type containing 18b |
| chr2_+_56534374 | 4.20 |
ENSDART00000113964
|
POLR2E (1 of many)
|
RNA polymerase II subunit E |
| chr11_-_18112090 | 4.16 |
ENSDART00000019248
|
tmem110
|
transmembrane protein 110 |
| chr2_-_9854212 | 4.13 |
ENSDART00000112995
|
wu:fi34b01
|
wu:fi34b01 |
| chr15_+_29091983 | 4.12 |
ENSDART00000141164
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr8_+_9660595 | 4.11 |
ENSDART00000091848
|
gripap1
|
GRIP1 associated protein 1 |
| chr1_+_47323244 | 4.11 |
|
|
|
| chr7_-_15008685 | 4.07 |
ENSDART00000173048
|
ENSDARG00000023868
|
ENSDARG00000023868 |
| chr19_+_14592882 | 4.07 |
ENSDART00000161088
ENSDART00000161965 |
zdhhc18b
|
zinc finger, DHHC-type containing 18b |
| chr22_+_24112851 | 4.07 |
|
|
|
| chr4_-_13615927 | 4.06 |
ENSDART00000138366
|
irf5
|
interferon regulatory factor 5 |
| chr5_-_64983760 | 4.04 |
ENSDART00000147707
|
camsap1b
|
calmodulin regulated spectrin-associated protein 1b |
| chr10_-_24350114 | 4.04 |
ENSDART00000109549
ENSDART00000148480 |
inpp5kb
|
inositol polyphosphate-5-phosphatase Kb |
| chr12_-_23244600 | 4.01 |
ENSDART00000170376
|
mpp7a
|
membrane protein, palmitoylated 7a (MAGUK p55 subfamily member 7) |
| chr22_+_17803347 | 4.01 |
ENSDART00000136016
|
hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr13_-_28480530 | 4.01 |
ENSDART00000043156
|
cyp17a1
|
cytochrome P450, family 17, subfamily A, polypeptide 1 |
| chr1_-_30289931 | 3.99 |
|
|
|
| chr20_-_14218080 | 3.98 |
ENSDART00000104032
|
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr14_-_32937341 | 3.96 |
ENSDART00000048130
|
stard14
|
START domain containing 14 |
| chr6_-_29314529 | 3.93 |
ENSDART00000132456
|
bivm
|
basic, immunoglobulin-like variable motif containing |
| chr7_+_69211965 | 3.89 |
ENSDART00000028064
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr1_-_30301568 | 3.85 |
ENSDART00000132466
|
si:ch211-269i23.2
|
si:ch211-269i23.2 |
| chr16_+_39209567 | 3.84 |
ENSDART00000121756
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr9_+_8418408 | 3.83 |
ENSDART00000043067
|
zgc:171776
|
zgc:171776 |
| chr14_-_7102535 | 3.83 |
ENSDART00000036463
|
dnd1
|
DND microRNA-mediated repression inhibitor 1 |
| chr13_-_37530730 | 3.82 |
ENSDART00000141295
|
si:dkey-188i13.11
|
si:dkey-188i13.11 |
| chr22_+_2237813 | 3.81 |
|
|
|
| chr24_+_20947881 | 3.78 |
ENSDART00000163621
|
ccdc191
|
coiled-coil domain containing 191 |
| chr16_+_46728897 | 3.73 |
ENSDART00000169767
|
rab25b
|
RAB25, member RAS oncogene family b |
| chr16_-_4350251 | 3.71 |
|
|
|
| chr22_-_26537845 | 3.69 |
|
|
|
| chr13_-_24538905 | 3.68 |
ENSDART00000000831
|
znf511
|
zinc finger protein 511 |
| chr20_-_6542402 | 3.67 |
ENSDART00000054653
|
mcm3l
|
MCM3 minichromosome maintenance deficient 3 (S. cerevisiae), like |
| chr15_+_38397897 | 3.65 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr11_+_30005716 | 3.64 |
ENSDART00000167618
|
CR790368.1
|
ENSDARG00000100936 |
| chr4_-_20456517 | 3.63 |
ENSDART00000003621
ENSDART00000132356 |
sinup
|
siaz-interacting nuclear protein |
| chr6_+_39186673 | 3.60 |
ENSDART00000156187
|
tac3b
|
tachykinin 3b |
| chr7_+_68964813 | 3.58 |
ENSDART00000166258
|
marveld3
|
MARVEL domain containing 3 |
| chr16_-_25317921 | 3.58 |
|
|
|
| chr5_-_21548930 | 3.56 |
ENSDART00000002938
|
mtmr8
|
myotubularin related protein 8 |
| chr23_-_32239909 | 3.56 |
|
|
|
| chr20_+_35305119 | 3.55 |
ENSDART00000045135
|
fbxo16
|
F-box protein 16 |
| chr15_+_12127861 | 3.54 |
|
|
|
| chr10_+_43953171 | 3.54 |
|
|
|
| chr5_-_64983812 | 3.54 |
ENSDART00000147707
|
camsap1b
|
calmodulin regulated spectrin-associated protein 1b |
| chr2_+_27730940 | 3.53 |
ENSDART00000134976
|
si:ch73-382f3.1
|
si:ch73-382f3.1 |
| chr13_-_36785584 | 3.53 |
ENSDART00000167154
|
trim9
|
tripartite motif containing 9 |
| chr7_-_24567184 | 3.53 |
ENSDART00000131530
|
fam113
|
family with sequence similarity 113 |
| chr10_-_1933874 | 3.51 |
ENSDART00000101023
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr13_-_9110887 | 3.50 |
ENSDART00000138833
|
grxcr1
|
glutaredoxin, cysteine rich 1 |
| chr17_-_36913302 | 3.49 |
ENSDART00000154981
|
senp6b
|
SUMO1/sentrin specific peptidase 6b |
| chr5_-_32636372 | 3.49 |
ENSDART00000085512
ENSDART00000144694 |
kank1b
|
KN motif and ankyrin repeat domains 1b |
| chr21_-_43671213 | 3.48 |
ENSDART00000139008
|
si:dkey-229d11.3
|
si:dkey-229d11.3 |
| chr5_-_64983964 | 3.42 |
ENSDART00000147707
|
camsap1b
|
calmodulin regulated spectrin-associated protein 1b |
| chr16_+_31871645 | 3.40 |
ENSDART00000045210
|
mlf2
|
myeloid leukemia factor 2 |
| chr8_+_2583789 | 3.40 |
ENSDART00000143242
|
naif1
|
nuclear apoptosis inducing factor 1 |
| chr23_-_18131502 | 3.40 |
ENSDART00000173075
|
zgc:92287
|
zgc:92287 |
| chr9_-_34459768 | 3.39 |
ENSDART00000059955
|
ildr1b
|
immunoglobulin-like domain containing receptor 1b |
| chr10_-_24350023 | 3.39 |
ENSDART00000109549
ENSDART00000148480 |
inpp5kb
|
inositol polyphosphate-5-phosphatase Kb |
| chr23_-_34017934 | 3.38 |
ENSDART00000133223
|
ENSDARG00000070393
|
ENSDARG00000070393 |
| chr5_-_13347964 | 3.36 |
ENSDART00000127109
|
figla
|
folliculogenesis specific bHLH transcription factor |
| chr21_-_43660946 | 3.36 |
ENSDART00000136392
|
si:ch211-263m18.4
|
si:ch211-263m18.4 |
| chr24_-_33894123 | 3.34 |
ENSDART00000128679
|
cdk5
|
cyclin-dependent kinase 5 |
| chr8_+_53173077 | 3.34 |
ENSDART00000131514
|
nadka
|
NAD kinase a |
| chr22_+_22413878 | 3.32 |
ENSDART00000147825
|
kif14
|
kinesin family member 14 |
| chr21_-_34227100 | 3.31 |
ENSDART00000124649
|
alg13
|
ALG13, UDP-N-acetylglucosaminyltransferase subunit |
| chr8_+_28715451 | 3.31 |
ENSDART00000140115
|
ccser1
|
coiled-coil serine-rich protein 1 |
| chr24_-_31967674 | 3.31 |
ENSDART00000156060
|
si:ch73-78o10.1
|
si:ch73-78o10.1 |
| chr18_-_25784843 | 3.29 |
ENSDART00000103046
|
zgc:162879
|
zgc:162879 |
| chr10_-_31861975 | 3.28 |
ENSDART00000077785
|
vps26bl
|
vacuolar protein sorting 26 homolog B, like |
| chr21_-_26379011 | 3.25 |
|
|
|
| chr7_+_19300487 | 3.24 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr5_-_56953716 | 3.22 |
ENSDART00000167892
|
zgc:193711
|
zgc:193711 |
| chr11_-_11353309 | 3.21 |
ENSDART00000016677
|
zgc:77929
|
zgc:77929 |
| chr6_-_12665211 | 3.21 |
ENSDART00000150887
|
ical1
|
islet cell autoantigen 1-like |
| chr2_-_2373601 | 3.21 |
ENSDART00000113774
|
si:ch211-188c16.1
|
si:ch211-188c16.1 |
| chr12_+_38632987 | 3.19 |
ENSDART00000155563
|
abca5
|
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr11_-_1524107 | 3.17 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr12_+_13704914 | 3.16 |
ENSDART00000152257
|
ppp1r16a
|
protein phosphatase 1, regulatory subunit 16A |
| chr5_-_56985835 | 3.16 |
|
|
|
| chr12_+_38633026 | 3.15 |
ENSDART00000155563
|
abca5
|
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr18_+_44775384 | 3.13 |
ENSDART00000016271
|
ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr16_-_22394669 | 3.13 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 14.5 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 3.6 | 3.6 | GO:0045777 | positive regulation of blood pressure(GO:0045777) |
| 3.5 | 20.9 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 2.8 | 8.5 | GO:0072045 | convergent extension involved in nephron morphogenesis(GO:0072045) |
| 2.8 | 8.3 | GO:2000725 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 2.6 | 10.6 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 2.6 | 26.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 2.3 | 9.2 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 2.0 | 10.1 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 1.8 | 7.4 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 1.8 | 5.4 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 1.8 | 12.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 1.6 | 27.4 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 1.4 | 7.0 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 1.4 | 9.7 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 1.4 | 18.0 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 1.4 | 30.1 | GO:0001840 | neural plate development(GO:0001840) |
| 1.3 | 5.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 1.3 | 5.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 1.3 | 5.1 | GO:0030238 | male sex determination(GO:0030238) androst-4-ene-3,17-dione biosynthetic process(GO:1903449) |
| 1.3 | 12.8 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 1.1 | 12.7 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) mitotic DNA replication(GO:1902969) |
| 1.1 | 10.6 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 1.0 | 6.2 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 1.0 | 11.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.9 | 8.5 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.9 | 2.6 | GO:0021755 | glial cell migration(GO:0008347) eurydendroid cell differentiation(GO:0021755) |
| 0.8 | 2.5 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.8 | 4.0 | GO:0008585 | female gonad development(GO:0008585) |
| 0.8 | 1.6 | GO:0021744 | medulla oblongata development(GO:0021550) dorsal motor nucleus of vagus nerve development(GO:0021744) |
| 0.8 | 1.6 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.8 | 3.8 | GO:0060967 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.7 | 3.0 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.7 | 2.9 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.7 | 0.7 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.7 | 3.4 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.7 | 2.7 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.7 | 2.0 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.6 | 1.9 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.6 | 20.9 | GO:0048599 | oocyte development(GO:0048599) |
| 0.6 | 4.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.6 | 6.1 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.6 | 1.8 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.6 | 1.2 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.6 | 2.3 | GO:0072387 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.6 | 2.3 | GO:0006407 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.6 | 3.3 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.6 | 7.2 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.5 | 4.8 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.5 | 9.5 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.5 | 17.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.5 | 2.1 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.5 | 1.5 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) cellular response to vitamin D(GO:0071305) |
| 0.5 | 2.0 | GO:0099558 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.5 | 3.0 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.5 | 4.0 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.5 | 3.5 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.5 | 10.6 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.5 | 0.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.4 | 3.5 | GO:0032196 | transposition(GO:0032196) |
| 0.4 | 4.4 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.4 | 5.6 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.4 | 7.2 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.4 | 70.8 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.4 | 10.9 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.4 | 1.6 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.4 | 2.2 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.4 | 3.2 | GO:0042542 | response to hydrogen peroxide(GO:0042542) |
| 0.4 | 1.8 | GO:0003379 | establishment of cell polarity involved in gastrulation cell migration(GO:0003379) |
| 0.4 | 1.1 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.3 | 5.9 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.3 | 3.8 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.3 | 4.7 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.3 | 1.9 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.3 | 2.5 | GO:0000022 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.3 | 7.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.3 | 3.1 | GO:0007525 | somatic muscle development(GO:0007525) muscle attachment(GO:0016203) |
| 0.3 | 8.7 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.3 | 1.5 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.3 | 0.6 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.3 | 1.1 | GO:1904353 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.3 | 2.2 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.3 | 1.6 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.3 | 1.3 | GO:0045981 | regulation of oxidative phosphorylation(GO:0002082) positive regulation of nucleotide metabolic process(GO:0045981) positive regulation of purine nucleotide metabolic process(GO:1900544) |
| 0.3 | 0.8 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.3 | 4.7 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.3 | 1.8 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.2 | 4.2 | GO:0010632 | regulation of epithelial cell migration(GO:0010632) |
| 0.2 | 2.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 6.0 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.2 | 11.6 | GO:0035304 | regulation of protein dephosphorylation(GO:0035304) |
| 0.2 | 1.1 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.2 | 3.2 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.2 | 0.9 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.2 | 1.3 | GO:0035264 | multicellular organism growth(GO:0035264) |
| 0.2 | 1.3 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.2 | 0.8 | GO:0032060 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.2 | 1.2 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.2 | 2.8 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 3.6 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.2 | 0.4 | GO:0071921 | regulation of sister chromatid cohesion(GO:0007063) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.2 | 0.8 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.2 | 0.6 | GO:0002030 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) positive regulation of calcium ion transport into cytosol(GO:0010524) positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) regulation of inhibitory G-protein coupled receptor phosphorylation(GO:1904323) positive regulation of inhibitory G-protein coupled receptor phosphorylation(GO:1904325) |
| 0.2 | 1.7 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.2 | 5.6 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.2 | 0.7 | GO:0042373 | vitamin K metabolic process(GO:0042373) cellular response to fluid shear stress(GO:0071498) |
| 0.2 | 1.1 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.2 | 1.6 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.2 | 1.9 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.2 | 1.1 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.2 | 0.9 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.2 | 1.2 | GO:2000095 | protein localization to ciliary transition zone(GO:1904491) regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.2 | 1.2 | GO:0048885 | neuromast deposition(GO:0048885) |
| 0.2 | 1.1 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.2 | 1.3 | GO:0070989 | oxidative demethylation(GO:0070989) |
| 0.2 | 2.9 | GO:0042407 | cristae formation(GO:0042407) |
| 0.2 | 11.4 | GO:0045765 | regulation of angiogenesis(GO:0045765) |
| 0.2 | 7.8 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 1.2 | GO:0006901 | vesicle coating(GO:0006901) |
| 0.1 | 0.6 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 1.7 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 7.8 | GO:0032272 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.1 | 1.5 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 4.1 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.1 | 10.9 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.1 | 4.3 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 1.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.1 | 2.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 1.1 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.3 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 1.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.6 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 2.5 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.1 | 0.2 | GO:0055016 | hypochord development(GO:0055016) |
| 0.1 | 1.3 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 0.7 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 4.9 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.1 | 0.8 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.1 | 1.4 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 0.7 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.3 | GO:0070198 | protein localization to chromosome, telomeric region(GO:0070198) |
| 0.1 | 1.3 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 0.3 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.1 | 0.5 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 5.1 | GO:0097191 | extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.1 | 0.6 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.1 | 1.6 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.1 | 0.5 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.1 | 7.2 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.1 | 0.7 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 1.7 | GO:0098661 | inorganic anion transmembrane transport(GO:0098661) |
| 0.1 | 0.8 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 0.3 | GO:2000623 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 6.7 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.1 | 0.6 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.3 | GO:1902414 | protein localization to adherens junction(GO:0071896) protein localization to cell junction(GO:1902414) |
| 0.1 | 0.5 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 1.9 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 6.0 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.1 | 12.2 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.1 | 4.9 | GO:0019216 | regulation of lipid metabolic process(GO:0019216) |
| 0.1 | 1.6 | GO:0071218 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.1 | 1.1 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.1 | 2.8 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.1 | 0.9 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.1 | 1.6 | GO:0007006 | mitochondrial membrane organization(GO:0007006) |
| 0.1 | 0.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 1.6 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.1 | 0.9 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 2.6 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.1 | 3.1 | GO:0000045 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.1 | 2.4 | GO:0051170 | nuclear import(GO:0051170) |
| 0.1 | 3.0 | GO:0006096 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.1 | 2.2 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 1.9 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.1 | 1.5 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.1 | 1.2 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.8 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 3.0 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 2.5 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.8 | GO:0051181 | cofactor transport(GO:0051181) |
| 0.0 | 1.2 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.2 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.0 | 1.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.7 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 1.0 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.6 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.2 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.6 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.8 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 1.2 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 1.5 | GO:0005977 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) energy reserve metabolic process(GO:0006112) glucan metabolic process(GO:0044042) |
| 0.0 | 1.7 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 0.7 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.3 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.0 | 5.3 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.1 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.2 | GO:0019673 | GDP-mannose biosynthetic process(GO:0009298) GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 2.0 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 1.5 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.9 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.8 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 15.2 | GO:0015031 | protein transport(GO:0015031) |
| 0.0 | 1.4 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.2 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 2.8 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.7 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 1.2 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.6 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.9 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.5 | GO:0048385 | regulation of retinoic acid receptor signaling pathway(GO:0048385) |
| 0.0 | 0.1 | GO:0061136 | regulation of proteasomal protein catabolic process(GO:0061136) regulation of proteolysis involved in cellular protein catabolic process(GO:1903050) |
| 0.0 | 1.4 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 1.8 | GO:0001947 | heart looping(GO:0001947) |
| 0.0 | 0.4 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 1.0 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 0.1 | GO:0034311 | diol metabolic process(GO:0034311) |
| 0.0 | 0.5 | GO:0000723 | telomere maintenance(GO:0000723) telomere organization(GO:0032200) |
| 0.0 | 6.8 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.3 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 1.9 | GO:0006511 | ubiquitin-dependent protein catabolic process(GO:0006511) |
| 0.0 | 0.3 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.5 | GO:0017156 | calcium ion regulated exocytosis(GO:0017156) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 14.5 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 3.9 | 11.7 | GO:0033391 | chromatoid body(GO:0033391) |
| 2.9 | 26.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 1.6 | 8.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 1.6 | 4.7 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 1.5 | 19.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 1.4 | 9.7 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 1.2 | 4.9 | GO:0044609 | DBIRD complex(GO:0044609) |
| 1.1 | 67.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 1.0 | 7.2 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.9 | 3.7 | GO:0098536 | deuterosome(GO:0098536) |
| 0.9 | 2.7 | GO:0000800 | lateral element(GO:0000800) |
| 0.8 | 8.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.8 | 4.7 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.7 | 6.1 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.7 | 3.4 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.7 | 12.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.6 | 2.2 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.5 | 6.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.5 | 1.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.4 | 7.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.4 | 6.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.3 | 3.8 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.3 | 1.6 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.3 | 3.1 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.3 | 3.0 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.3 | 9.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.3 | 15.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.3 | 1.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.3 | 1.9 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.3 | 1.3 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.3 | 0.8 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.3 | 4.1 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.2 | 3.0 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 0.7 | GO:1905202 | methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.2 | 2.5 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.2 | 2.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.2 | 1.6 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.2 | 4.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 4.1 | GO:0043186 | P granule(GO:0043186) |
| 0.2 | 19.9 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.2 | 7.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 10.5 | GO:0001726 | ruffle(GO:0001726) |
| 0.2 | 1.0 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.2 | 2.7 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.2 | 2.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.2 | 0.8 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 1.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 6.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 11.7 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 2.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 5.0 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 0.7 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 1.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.9 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 6.3 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 11.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 6.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 0.8 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 4.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 1.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 1.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 2.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 15.6 | GO:0005938 | cell cortex(GO:0005938) |
| 0.1 | 6.5 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.1 | 0.2 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.1 | 2.8 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 1.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 1.3 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.5 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 1.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 0.7 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 0.6 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 5.3 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 0.3 | GO:0017054 | negative cofactor 2 complex(GO:0017054) |
| 0.1 | 0.6 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 0.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 4.3 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.2 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.9 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 5.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 3.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.5 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 3.5 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 1.5 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.5 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 10.1 | GO:0030529 | intracellular ribonucleoprotein complex(GO:0030529) |
| 0.0 | 2.3 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 5.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 5.4 | GO:0005794 | Golgi apparatus(GO:0005794) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 11.0 | GO:0004937 | alpha-adrenergic receptor activity(GO:0004936) alpha1-adrenergic receptor activity(GO:0004937) |
| 2.6 | 10.6 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 2.5 | 10.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 2.3 | 9.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 1.8 | 12.5 | GO:0035173 | histone kinase activity(GO:0035173) |
| 1.7 | 26.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 1.7 | 6.8 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 1.7 | 15.1 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 1.5 | 12.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 1.5 | 10.6 | GO:0043495 | protein anchor(GO:0043495) |
| 1.3 | 5.1 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 1.3 | 5.1 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 1.2 | 67.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 1.2 | 7.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 1.1 | 3.4 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 1.0 | 9.2 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.9 | 10.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.9 | 2.7 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.9 | 2.6 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.8 | 3.2 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.8 | 8.5 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.8 | 14.0 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.7 | 2.9 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.7 | 11.5 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.7 | 2.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.7 | 17.2 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.6 | 3.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.6 | 2.3 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.6 | 16.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.5 | 2.6 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.5 | 3.0 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.5 | 7.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.5 | 2.0 | GO:0051978 | lysophospholipid transporter activity(GO:0051978) |
| 0.5 | 1.5 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.5 | 1.4 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.5 | 1.9 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.4 | 2.7 | GO:0043531 | ADP binding(GO:0043531) |
| 0.4 | 3.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.4 | 4.7 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.4 | 17.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.4 | 10.9 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.4 | 2.4 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.4 | 1.6 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.4 | 1.4 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.3 | 7.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.3 | 4.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.3 | 1.3 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.3 | 1.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.3 | 2.9 | GO:0051429 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.3 | 7.7 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.3 | 6.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 2.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.3 | 1.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.3 | 3.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.3 | 4.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.3 | 9.4 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.3 | 0.9 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.3 | 6.0 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.3 | 1.3 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.3 | 1.1 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.3 | 5.3 | GO:0015296 | anion:cation symporter activity(GO:0015296) |
| 0.3 | 0.8 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.2 | 1.9 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.2 | 0.7 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.2 | 1.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.2 | 2.8 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.2 | 2.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 3.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 3.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 2.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 3.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.2 | 2.0 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.2 | 3.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.2 | 0.7 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.2 | 2.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.2 | 1.7 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.2 | 0.8 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.2 | 3.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 0.8 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.2 | 5.3 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.2 | 0.6 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.1 | 2.0 | GO:0015368 | calcium:cation antiporter activity(GO:0015368) |
| 0.1 | 1.1 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 0.5 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.1 | 2.9 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 1.8 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 0.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 1.3 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 2.0 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 1.2 | GO:0031995 | fibronectin binding(GO:0001968) insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.9 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.6 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 2.9 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.1 | 1.5 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 0.5 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.6 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 16.4 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 1.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 17.8 | GO:0008234 | cysteine-type peptidase activity(GO:0008234) |
| 0.1 | 1.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 9.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 1.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.6 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.1 | 1.8 | GO:0048038 | quinone binding(GO:0048038) |
| 0.1 | 2.8 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.1 | 0.5 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 10.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 0.6 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.1 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.8 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 1.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 4.9 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 1.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.7 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.9 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 0.7 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 2.2 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 2.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 3.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 2.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 2.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 1.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 1.0 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.2 | GO:0004475 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.1 | 1.1 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 1.2 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 4.3 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.1 | 0.2 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.1 | 1.5 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 0.3 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 1.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 3.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 3.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 12.0 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.6 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.7 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 18.9 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 1.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0001047 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.0 | 3.2 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 14.2 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.2 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.7 | GO:0032934 | sterol binding(GO:0032934) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.6 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 6.3 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.2 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.3 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.9 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.2 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 6.0 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 3.2 | GO:0016791 | phosphatase activity(GO:0016791) |
| 0.0 | 2.2 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.4 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 1.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 12.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.6 | 16.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.6 | 8.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.5 | 11.9 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.5 | 8.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.4 | 3.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.3 | 7.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.3 | 4.9 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.3 | 2.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 8.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.2 | 2.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 1.9 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 3.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 0.7 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 1.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 4.0 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 4.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.7 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.3 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 4.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.0 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.7 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 1.8 | 12.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 1.2 | 8.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 1.1 | 3.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.7 | 12.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.6 | 5.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.5 | 18.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.5 | 11.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.3 | 6.0 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.3 | 2.4 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.3 | 1.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.3 | 2.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 5.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.2 | 4.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.2 | 1.6 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.2 | 1.8 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.2 | 0.7 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 0.6 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.2 | 0.8 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.2 | 2.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 6.3 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.1 | 4.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 2.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 0.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 0.8 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 0.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.1 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 1.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 2.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 8.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.6 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.7 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.2 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.9 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.0 | 0.1 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.3 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |