Project
DANIO-CODE
Navigation
Downloads
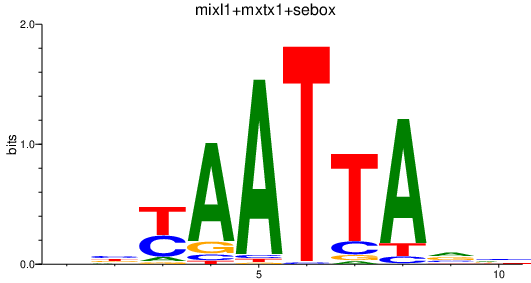
Results for mixl1+mxtx1+sebox
Z-value: 0.81
Transcription factors associated with mixl1+mxtx1+sebox
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sebox
|
ENSDARG00000042526 | SEBOX homeobox |
|
mixl1
|
ENSDARG00000069252 | Mix paired-like homeobox |
|
mxtx1
|
ENSDARG00000069382 | mix-type homeobox gene 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mxtx1 | dr10_dc_chr13_-_21529695_21529754 | -0.30 | 2.7e-01 | Click! |
| mixl1 | dr10_dc_chr20_-_43826581_43826593 | 0.27 | 3.1e-01 | Click! |
| sebox | dr10_dc_chr5_+_66693263_66693284 | -0.12 | 6.7e-01 | Click! |
Activity profile of mixl1+mxtx1+sebox motif
Sorted Z-values of mixl1+mxtx1+sebox motif
Network of associatons between targets according to the STRING database.
First level regulatory network of mixl1+mxtx1+sebox
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_25740782 | 4.97 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr18_-_40718244 | 3.29 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr10_-_34971985 | 3.06 |
ENSDART00000141201
|
ccna1
|
cyclin A1 |
| chr8_+_45326435 | 2.99 |
ENSDART00000134161
|
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr12_-_14104939 | 2.91 |
ENSDART00000152742
|
buc2l
|
bucky ball 2-like |
| chr17_+_16038358 | 2.73 |
ENSDART00000155336
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr24_+_12689711 | 2.61 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr11_-_6442588 | 2.58 |
ENSDART00000137879
|
zgc:162969
|
zgc:162969 |
| chr10_-_25246786 | 2.58 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr1_-_18118467 | 2.53 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr5_+_37303599 | 2.44 |
ENSDART00000097754
ENSDART00000162470 |
tmprss4b
|
transmembrane protease, serine 4b |
| chr10_-_21404605 | 2.44 |
ENSDART00000125167
|
avd
|
avidin |
| chr17_-_45021393 | 2.42 |
|
|
|
| chr14_+_34150130 | 2.31 |
ENSDART00000132193
ENSDART00000141058 |
wnt8a
BX927327.1
|
wingless-type MMTV integration site family, member 8a ENSDARG00000105311 |
| chr20_-_14218080 | 2.26 |
ENSDART00000104032
|
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr16_-_29452509 | 2.16 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr22_-_17627900 | 2.16 |
ENSDART00000138483
|
si:ch73-243b8.4
|
si:ch73-243b8.4 |
| chr19_+_15536640 | 2.07 |
ENSDART00000098970
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr17_+_16038103 | 2.04 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr1_-_54570813 | 2.03 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr20_-_23527234 | 1.97 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr24_+_8702288 | 1.95 |
ENSDART00000114810
|
sycp2l
|
synaptonemal complex protein 2-like |
| chr10_-_34971926 | 1.90 |
ENSDART00000141201
|
ccna1
|
cyclin A1 |
| chr2_+_6341404 | 1.78 |
ENSDART00000076700
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr21_-_32027717 | 1.77 |
ENSDART00000131651
|
ENSDARG00000073961
|
ENSDARG00000073961 |
| chr24_+_19270877 | 1.66 |
|
|
|
| chr16_+_47283253 | 1.65 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr10_+_6925373 | 1.65 |
ENSDART00000128866
|
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr3_+_28729443 | 1.56 |
ENSDART00000077235
|
alg1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr20_+_14218237 | 1.50 |
ENSDART00000044937
|
kcns3b
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3b |
| chr22_+_4035577 | 1.49 |
ENSDART00000170620
|
ctxn1
|
cortexin 1 |
| chr24_+_12689887 | 1.45 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr14_-_8634381 | 1.36 |
ENSDART00000129030
|
zgc:153681
|
zgc:153681 |
| KN149710v1_+_38638 | 1.32 |
|
|
|
| chr1_+_18118735 | 1.30 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr16_+_47283374 | 1.26 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr18_-_43890836 | 1.23 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr10_-_25448712 | 1.22 |
ENSDART00000140023
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr18_+_20571460 | 1.20 |
ENSDART00000151990
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr16_-_29451902 | 1.18 |
|
|
|
| chr1_+_21244200 | 1.16 |
ENSDART00000087729
|
kdm4c
|
lysine (K)-specific demethylase 4C |
| chr1_+_35253862 | 1.16 |
ENSDART00000139636
|
zgc:152968
|
zgc:152968 |
| chr10_-_32550351 | 1.14 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| chr6_+_40925259 | 1.12 |
ENSDART00000002728
ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr19_-_5186692 | 1.12 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr11_+_34909167 | 1.09 |
ENSDART00000110839
|
mon1a
|
MON1 secretory trafficking family member A |
| chr8_+_41003546 | 1.08 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr11_-_34909095 | 1.08 |
ENSDART00000017393
|
traip
|
TRAF-interacting protein |
| chr11_+_34909244 | 1.07 |
ENSDART00000110839
|
mon1a
|
MON1 secretory trafficking family member A |
| chr8_+_11387135 | 1.04 |
|
|
|
| chr15_-_16241412 | 1.03 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr8_-_44247277 | 1.02 |
|
|
|
| chr16_+_42567707 | 1.02 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr14_+_23420053 | 1.01 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr18_+_20571400 | 0.99 |
ENSDART00000151990
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr7_+_28341426 | 0.99 |
ENSDART00000019991
|
slc7a6os
|
solute carrier family 7, member 6 opposite strand |
| chr21_+_34053739 | 0.96 |
ENSDART00000147519
|
mtmr1b
|
myotubularin related protein 1b |
| chr17_-_25630635 | 0.96 |
ENSDART00000149060
|
ppp1cb
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr2_-_27892824 | 0.92 |
|
|
|
| chr5_+_6391432 | 0.83 |
ENSDART00000170564
ENSDART00000086666 |
stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr20_-_14218236 | 0.81 |
ENSDART00000168434
|
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr24_+_16924542 | 0.80 |
ENSDART00000014787
|
pip4k2aa
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha a |
| chr20_+_54404987 | 0.79 |
ENSDART00000099338
|
actr10
|
ARP10 actin related protein 10 homolog |
| chr1_+_21244242 | 0.77 |
ENSDART00000087729
|
kdm4c
|
lysine (K)-specific demethylase 4C |
| chr8_-_1833095 | 0.75 |
ENSDART00000114476
ENSDART00000091235 ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr21_+_34053590 | 0.74 |
ENSDART00000147519
ENSDART00000158115 ENSDART00000029599 ENSDART00000145123 |
mtmr1b
|
myotubularin related protein 1b |
| chr18_+_19467527 | 0.74 |
ENSDART00000079695
|
zwilch
|
zwilch kinetochore protein |
| chr8_-_19236074 | 0.74 |
ENSDART00000137994
|
zgc:77486
|
zgc:77486 |
| chr20_-_16271738 | 0.72 |
ENSDART00000012476
|
coa7
|
cytochrome c oxidase assembly factor 7 |
| chr21_-_36710989 | 0.71 |
ENSDART00000086060
|
mrpl22
|
mitochondrial ribosomal protein L22 |
| chr11_-_6442490 | 0.69 |
ENSDART00000137879
|
zgc:162969
|
zgc:162969 |
| chr6_+_48188549 | 0.69 |
|
|
|
| chr23_-_25208472 | 0.67 |
ENSDART00000103989
ENSDART00000160278 |
idh3g
|
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr21_-_25765319 | 0.66 |
ENSDART00000101219
|
mettl27
|
methyltransferase like 27 |
| chr1_+_8010026 | 0.66 |
ENSDART00000152142
|
zgc:77849
|
zgc:77849 |
| chr20_-_37910887 | 0.66 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr2_+_6341345 | 0.65 |
ENSDART00000058256
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr5_-_18459312 | 0.64 |
ENSDART00000145210
|
ankle2
|
ankyrin repeat and LEM domain containing 2 |
| chr14_+_23419864 | 0.63 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr6_+_50382153 | 0.63 |
ENSDART00000055504
|
cyc1
|
cytochrome c-1 |
| chr7_+_26378093 | 0.63 |
ENSDART00000173823
ENSDART00000101053 |
tp53i11a
|
tumor protein p53 inducible protein 11a |
| chr3_-_39554099 | 0.62 |
ENSDART00000145303
|
b9d1
|
B9 protein domain 1 |
| chr8_-_21039978 | 0.61 |
ENSDART00000137606
ENSDART00000146532 |
zgc:112962
|
zgc:112962 |
| chr11_-_12024136 | 0.61 |
ENSDART00000111919
|
sp2
|
sp2 transcription factor |
| chr18_+_20571785 | 0.60 |
ENSDART00000040074
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr14_-_33605295 | 0.59 |
ENSDART00000168546
|
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr7_+_8492338 | 0.58 |
|
|
|
| chr13_-_35346618 | 0.58 |
ENSDART00000057052
|
slx4ip
|
SLX4 interacting protein |
| chr23_-_31986679 | 0.56 |
ENSDART00000085054
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr5_+_37303707 | 0.54 |
ENSDART00000097754
ENSDART00000162470 |
tmprss4b
|
transmembrane protease, serine 4b |
| chr8_+_45326523 | 0.54 |
ENSDART00000145011
|
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr12_+_3632548 | 0.54 |
|
|
|
| chr4_-_5822882 | 0.54 |
ENSDART00000008898
|
foxm1
|
forkhead box M1 |
| chr11_+_34909393 | 0.53 |
ENSDART00000110839
|
mon1a
|
MON1 secretory trafficking family member A |
| chr3_-_26112995 | 0.53 |
ENSDART00000103741
|
ppp4ca
|
protein phosphatase 4, catalytic subunit a |
| chr8_+_50964745 | 0.52 |
ENSDART00000013870
|
ENSDARG00000007359
|
ENSDARG00000007359 |
| chr3_-_26113336 | 0.51 |
ENSDART00000103741
|
ppp4ca
|
protein phosphatase 4, catalytic subunit a |
| chr3_+_26113651 | 0.51 |
ENSDART00000103734
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr14_+_34150232 | 0.51 |
ENSDART00000148044
|
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| KN150200v1_+_7782 | 0.49 |
|
|
|
| chr6_-_32001100 | 0.49 |
ENSDART00000132280
|
ror1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr8_-_5834829 | 0.48 |
ENSDART00000179217
|
ENSDARG00000106522
|
ENSDARG00000106522 |
| chr24_+_16402587 | 0.48 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr24_-_24999348 | 0.48 |
ENSDART00000152104
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr8_+_36467811 | 0.47 |
ENSDART00000098701
|
slc7a4
|
solute carrier family 7, member 4 |
| chr15_-_16241500 | 0.46 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr24_-_14447825 | 0.45 |
|
|
|
| chr25_-_28630138 | 0.45 |
|
|
|
| chr11_+_26366532 | 0.45 |
ENSDART00000159505
|
dynlrb1
|
dynein, light chain, roadblock-type 1 |
| chr6_+_49489473 | 0.43 |
ENSDART00000177749
|
FO704848.1
|
ENSDARG00000107438 |
| chr16_+_42567668 | 0.43 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr8_-_25015215 | 0.42 |
ENSDART00000170511
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr16_-_42105636 | 0.41 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr3_+_26113393 | 0.41 |
ENSDART00000103733
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr14_+_8634323 | 0.41 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr8_+_26377369 | 0.39 |
ENSDART00000087151
|
amt
|
aminomethyltransferase |
| chr6_+_48188609 | 0.37 |
|
|
|
| chr24_-_14447655 | 0.36 |
|
|
|
| chr6_-_51771558 | 0.35 |
ENSDART00000073847
|
blcap
|
bladder cancer associated protein |
| chr8_+_41003629 | 0.35 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr17_-_41000670 | 0.34 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr24_+_12689972 | 0.34 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr17_+_8642373 | 0.34 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr23_-_31986571 | 0.33 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr24_-_14447519 | 0.32 |
|
|
|
| chr22_-_20141216 | 0.28 |
ENSDART00000128023
|
btbd2a
|
BTB (POZ) domain containing 2a |
| chr11_+_33555374 | 0.26 |
ENSDART00000109418
|
spoplb
|
speckle-type POZ protein-like b |
| chr16_-_42105733 | 0.26 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr25_-_24440292 | 0.25 |
ENSDART00000156805
|
CR854832.1
|
ENSDARG00000096817 |
| chr17_-_41809859 | 0.24 |
ENSDART00000156031
|
ralgapa2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
| chr23_+_22763739 | 0.22 |
|
|
|
| chr3_-_13311188 | 0.21 |
ENSDART00000080807
|
fbxw9
|
F-box and WD repeat domain containing 9 |
| chr11_+_34259003 | 0.21 |
|
|
|
| chr24_-_24578984 | 0.21 |
ENSDART00000012399
|
armc1
|
armadillo repeat containing 1 |
| chr10_+_6925975 | 0.21 |
|
|
|
| chr24_+_12690117 | 0.19 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr22_+_20141311 | 0.18 |
|
|
|
| chr16_-_26798351 | 0.18 |
|
|
|
| chr15_-_45304762 | 0.15 |
ENSDART00000177018
|
CABZ01068243.1
|
ENSDARG00000106883 |
| chr17_+_8642428 | 0.15 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr5_-_66441778 | 0.14 |
ENSDART00000168245
|
rps6ka4
|
ribosomal protein S6 kinase, polypeptide 4 |
| chr24_+_16924429 | 0.12 |
ENSDART00000014787
|
pip4k2aa
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha a |
| chr2_+_50873843 | 0.10 |
|
|
|
| chr23_-_31986482 | 0.08 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr13_+_25590508 | 0.07 |
ENSDART00000046050
|
pcbd1
|
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha |
| chr1_+_35253821 | 0.07 |
ENSDART00000179634
|
zgc:152968
|
zgc:152968 |
| chr8_-_16370646 | 0.06 |
ENSDART00000115419
|
faf1
|
Fas (TNFRSF6) associated factor 1 |
| chr20_-_21031592 | 0.03 |
ENSDART00000152726
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr1_-_28335293 | 0.03 |
ENSDART00000075546
|
hsf2bp
|
heat shock transcription factor 2 binding protein |
| chr16_+_6809981 | 0.02 |
ENSDART00000149720
|
znf236
|
zinc finger protein 236 |
| chr15_+_1569864 | 0.02 |
ENSDART00000056765
|
smc4
|
structural maintenance of chromosomes 4 |
| chr15_-_16241341 | 0.01 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr11_-_36188531 | 0.01 |
ENSDART00000128889
|
zbtb40
|
zinc finger and BTB domain containing 40 |
| chr14_+_23419894 | 0.00 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.6 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.7 | 2.6 | GO:0039015 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.6 | 2.8 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.5 | 1.4 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.4 | 1.1 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.2 | 2.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 2.9 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.2 | 5.0 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.2 | 0.7 | GO:0030730 | regulation of sequestering of triglyceride(GO:0010889) positive regulation of sequestering of triglyceride(GO:0010890) sequestering of triglyceride(GO:0030730) |
| 0.2 | 3.1 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 1.7 | GO:0061458 | gonad development(GO:0008406) reproductive structure development(GO:0048608) reproductive system development(GO:0061458) |
| 0.2 | 0.8 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 0.7 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.1 | 1.0 | GO:0048532 | anatomical structure arrangement(GO:0048532) |
| 0.1 | 0.6 | GO:0035264 | multicellular organism growth(GO:0035264) |
| 0.1 | 0.4 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.7 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 5.0 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 0.6 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 2.7 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.1 | 0.6 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.6 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.5 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.1 | 2.0 | GO:0048599 | oocyte development(GO:0048599) |
| 0.0 | 1.2 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 1.0 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 1.0 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 1.6 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 1.7 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.4 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 2.2 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 0.5 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 1.5 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 2.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.5 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 0.6 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.6 | GO:0048634 | regulation of muscle organ development(GO:0048634) |
| 0.0 | 0.6 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.7 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 1.8 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.3 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) negative regulation of protein modification by small protein conjugation or removal(GO:1903321) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.7 | 2.6 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.3 | 2.9 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.2 | 3.7 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 4.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 1.5 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 0.5 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 0.6 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.8 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 4.5 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 3.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.7 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.7 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 1.2 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.4 | GO:0009374 | biotin binding(GO:0009374) |
| 0.4 | 1.1 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.3 | 1.4 | GO:1903231 | mRNA binding involved in posttranscriptional gene silencing(GO:1903231) |
| 0.3 | 3.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.3 | 2.4 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.2 | 1.4 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.2 | 1.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 1.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 1.8 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.2 | 0.7 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.2 | 2.6 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 0.8 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 1.2 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 3.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.7 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 5.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 2.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.6 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 2.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 1.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 2.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 1.5 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 9.6 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.6 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.5 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 2.1 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 2.9 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 1.2 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.6 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 0.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 4.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.5 | ST GA13 PATHWAY | G alpha 13 Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.0 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |