Project
DANIO-CODE
Navigation
Downloads
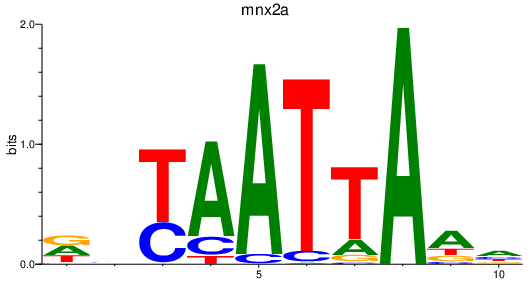
Results for mnx2a
Z-value: 0.46
Transcription factors associated with mnx2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mnx2a
|
ENSDARG00000042106 | motor neuron and pancreas homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mnx2a | dr10_dc_chr9_+_7745593_7745601 | 0.62 | 1.1e-02 | Click! |
Activity profile of mnx2a motif
Sorted Z-values of mnx2a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of mnx2a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_56157608 | 1.49 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr18_-_3056732 | 1.17 |
ENSDART00000162657
|
rps3
|
ribosomal protein S3 |
| chr16_+_23516127 | 1.10 |
ENSDART00000004679
|
icn
|
ictacalcin |
| chr17_+_23278879 | 0.93 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr20_-_22576513 | 0.92 |
ENSDART00000103510
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr22_-_26333957 | 0.89 |
ENSDART00000130493
|
capn2b
|
calpain 2, (m/II) large subunit b |
| chr3_-_29768364 | 0.72 |
ENSDART00000020311
|
rpl27
|
ribosomal protein L27 |
| chr3_+_25992836 | 0.63 |
ENSDART00000010477
|
hsp70.3
|
heat shock cognate 70-kd protein, tandem duplicate 3 |
| chr12_+_24221087 | 0.63 |
ENSDART00000088178
|
nrxn1a
|
neurexin 1a |
| chr7_-_25623974 | 0.61 |
ENSDART00000173602
|
cd99l2
|
CD99 molecule-like 2 |
| chr19_-_43037791 | 0.61 |
ENSDART00000004392
|
fkbp9
|
FK506 binding protein 9 |
| chr9_-_31467299 | 0.60 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr14_-_17257773 | 0.58 |
ENSDART00000082667
|
fgfrl1a
|
fibroblast growth factor receptor-like 1a |
| chr23_+_25428372 | 0.57 |
ENSDART00000147440
|
fmnl3
|
formin-like 3 |
| chr15_-_4537178 | 0.57 |
ENSDART00000155619
ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr3_-_27516974 | 0.56 |
ENSDART00000151675
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr24_-_6048914 | 0.54 |
ENSDART00000146830
ENSDART00000021981 |
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr2_-_34010299 | 0.53 |
ENSDART00000140910
|
ptch2
|
patched 2 |
| chr25_-_19388415 | 0.51 |
ENSDART00000156016
|
zgc:193812
|
zgc:193812 |
| chr2_+_49358871 | 0.50 |
ENSDART00000179089
|
sema6ba
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Ba |
| chr21_+_28441951 | 0.49 |
ENSDART00000077887
|
slc22a6l
|
solute carrier family 22 (organic anion transporter), member 6, like |
| chr8_-_34077387 | 0.49 |
ENSDART00000159208
ENSDART00000040126 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr14_+_20045365 | 0.48 |
ENSDART00000167637
|
aff2
|
AF4/FMR2 family, member 2 |
| chr15_-_23441268 | 0.48 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr9_-_20562293 | 0.45 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr16_-_55320569 | 0.45 |
ENSDART00000156368
|
ENSDARG00000069583
|
ENSDARG00000069583 |
| chr6_+_23787804 | 0.45 |
ENSDART00000163188
|
znf648
|
zinc finger protein 648 |
| chr16_+_23172295 | 0.44 |
ENSDART00000167518
ENSDART00000161087 |
efna3b
|
ephrin-A3b |
| chr14_+_160149 | 0.44 |
ENSDART00000158405
ENSDART00000158072 |
gpc2
|
glypican 2 |
| chr5_-_41672394 | 0.43 |
ENSDART00000164363
|
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr14_+_36156947 | 0.42 |
|
|
|
| chr19_-_6384776 | 0.42 |
|
|
|
| chr14_+_21816442 | 0.41 |
ENSDART00000109759
|
tmx2a
|
thioredoxin-related transmembrane protein 2a |
| chr20_-_9107294 | 0.41 |
ENSDART00000140792
|
OMA1
|
OMA1 zinc metallopeptidase |
| chr3_-_6078015 | 0.40 |
ENSDART00000165715
|
BX284638.1
|
ENSDARG00000098850 |
| chr23_-_31585883 | 0.39 |
ENSDART00000157511
|
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr24_+_21395671 | 0.39 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr1_-_25600988 | 0.38 |
ENSDART00000160381
|
cxxc4
|
CXXC finger 4 |
| chr14_-_2682064 | 0.38 |
ENSDART00000161677
|
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr22_+_2921305 | 0.38 |
ENSDART00000143258
|
cep19
|
centrosomal protein 19 |
| chr20_+_21002097 | 0.37 |
ENSDART00000035827
|
brf1b
|
BRF1, RNA polymerase III transcription initiation factor b |
| chr8_+_39964695 | 0.37 |
ENSDART00000073782
|
ggt5a
|
gamma-glutamyltransferase 5a |
| chr22_+_16509286 | 0.37 |
ENSDART00000083063
|
tal1
|
T-cell acute lymphocytic leukemia 1 |
| chr22_+_12746080 | 0.36 |
ENSDART00000044683
|
ftcd
|
formimidoyltransferase cyclodeaminase |
| chr20_-_48677794 | 0.36 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr17_-_52735250 | 0.36 |
|
|
|
| chr2_-_40170303 | 0.36 |
ENSDART00000165602
|
epha4a
|
eph receptor A4a |
| chr22_-_20141589 | 0.36 |
ENSDART00000085913
|
btbd2a
|
BTB (POZ) domain containing 2a |
| chr4_+_16896821 | 0.36 |
ENSDART00000017726
|
etnk1
|
ethanolamine kinase 1 |
| chr6_+_24299180 | 0.34 |
ENSDART00000167482
|
tgfbr3
|
transforming growth factor, beta receptor III |
| chr16_+_51319421 | 0.34 |
|
|
|
| chr16_+_21436660 | 0.34 |
ENSDART00000145886
|
osbpl3b
|
oxysterol binding protein-like 3b |
| chr18_-_14709371 | 0.33 |
ENSDART00000111995
|
si:dkey-238o13.4
|
si:dkey-238o13.4 |
| chr17_+_30352361 | 0.33 |
ENSDART00000076611
|
greb1
|
growth regulation by estrogen in breast cancer 1 |
| chr16_-_21238288 | 0.33 |
ENSDART00000139737
|
cbx3b
|
chromobox homolog 3b |
| chr1_-_40208544 | 0.33 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr6_+_13833991 | 0.32 |
|
|
|
| chr4_+_3970874 | 0.32 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr1_-_40208469 | 0.32 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr24_-_14447773 | 0.31 |
|
|
|
| chr25_+_30701751 | 0.31 |
|
|
|
| chr14_+_4169846 | 0.30 |
ENSDART00000038301
|
gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr11_-_6051096 | 0.30 |
ENSDART00000147761
|
vsg1
|
vessel-specific 1 |
| chr8_-_11512545 | 0.30 |
ENSDART00000133932
|
si:ch211-248e11.2
|
si:ch211-248e11.2 |
| chr19_-_43038013 | 0.30 |
ENSDART00000004392
|
fkbp9
|
FK506 binding protein 9 |
| chr12_+_5046825 | 0.29 |
ENSDART00000164178
|
prrt2
|
proline-rich transmembrane protein 2 |
| chr19_+_5562107 | 0.29 |
ENSDART00000082080
|
jupb
|
junction plakoglobin b |
| chr6_+_45917081 | 0.28 |
ENSDART00000149450
ENSDART00000149642 |
h6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr20_+_28958586 | 0.28 |
ENSDART00000142678
|
slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr11_-_6442836 | 0.28 |
ENSDART00000004483
|
zgc:162969
|
zgc:162969 |
| chr19_+_22478256 | 0.27 |
ENSDART00000100181
|
sall3b
|
spalt-like transcription factor 3b |
| chr8_-_19166630 | 0.26 |
|
|
|
| chr18_-_43890514 | 0.26 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr21_+_26684617 | 0.25 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr22_-_20141216 | 0.25 |
ENSDART00000128023
|
btbd2a
|
BTB (POZ) domain containing 2a |
| chr23_+_36016450 | 0.25 |
ENSDART00000103035
|
hoxc6a
|
homeobox C6a |
| chr9_-_30243398 | 0.25 |
ENSDART00000139863
ENSDART00000140040 |
dcbld2
|
discoidin, CUB and LCCL domain containing 2 |
| chr3_+_27667194 | 0.25 |
ENSDART00000075100
|
carhsp1
|
calcium regulated heat stable protein 1 |
| chr7_+_25052687 | 0.25 |
ENSDART00000110347
|
cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr13_+_25590508 | 0.24 |
ENSDART00000046050
|
pcbd1
|
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha |
| chr10_-_24401876 | 0.24 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr3_-_27517052 | 0.24 |
ENSDART00000151625
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| KN149726v1_+_1094 | 0.24 |
|
|
|
| chr13_-_44492897 | 0.23 |
|
|
|
| chr8_+_2428689 | 0.23 |
ENSDART00000081325
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr1_+_11290598 | 0.22 |
ENSDART00000103399
|
tspan5b
|
tetraspanin 5b |
| chr25_+_15842875 | 0.22 |
ENSDART00000126641
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr17_-_52735615 | 0.22 |
|
|
|
| chr8_+_43334181 | 0.22 |
ENSDART00000038566
|
fam101a
|
family with sequence similarity 101, member A |
| chr7_+_19300351 | 0.21 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr22_-_2921219 | 0.21 |
ENSDART00000092991
|
pigx
|
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr9_+_34832049 | 0.21 |
ENSDART00000100735
ENSDART00000133996 |
shox
|
short stature homeobox |
| chr1_-_50215233 | 0.20 |
ENSDART00000137648
|
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr6_-_57542101 | 0.19 |
ENSDART00000156967
|
itcha
|
itchy E3 ubiquitin protein ligase a |
| chr17_-_37447917 | 0.19 |
ENSDART00000075975
|
crip1
|
cysteine-rich protein 1 |
| chr18_-_16600954 | 0.19 |
ENSDART00000143744
|
mgat4c
|
mgat4 family, member C |
| chr10_+_37193690 | 0.19 |
ENSDART00000114909
|
cuedc1a
|
CUE domain containing 1a |
| chr7_-_25624212 | 0.18 |
ENSDART00000173505
|
cd99l2
|
CD99 molecule-like 2 |
| chr1_-_31379323 | 0.18 |
ENSDART00000101958
|
pnpla4
|
patatin-like phospholipase domain containing 4 |
| chr23_+_17220363 | 0.18 |
ENSDART00000143420
|
BX927275.2
|
ENSDARG00000095017 |
| chr14_-_15845445 | 0.17 |
ENSDART00000162431
|
ptcd3
|
pentatricopeptide repeat domain 3 |
| chr6_-_11544518 | 0.17 |
ENSDART00000151195
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr14_-_32543646 | 0.17 |
ENSDART00000105726
|
slc25a5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr24_-_6641762 | 0.17 |
ENSDART00000114700
|
unm_sa821
|
un-named sa821 |
| chr17_-_37447869 | 0.17 |
ENSDART00000148160
|
crip1
|
cysteine-rich protein 1 |
| chr8_+_25015325 | 0.17 |
ENSDART00000140617
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr20_-_37587804 | 0.17 |
ENSDART00000166106
|
si:ch211-202p1.5
|
si:ch211-202p1.5 |
| chr11_-_19612693 | 0.16 |
ENSDART00000037894
|
namptb
|
nicotinamide phosphoribosyltransferase b |
| chr6_+_28215039 | 0.16 |
ENSDART00000104394
|
smx5
|
smx5 |
| chr15_-_16241341 | 0.16 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr23_-_40873523 | 0.16 |
ENSDART00000127420
|
ENSDARG00000088040
|
ENSDARG00000088040 |
| chr13_+_47175 | 0.15 |
ENSDART00000102505
|
foxg1d
|
forkhead box G1d |
| chr19_+_42899678 | 0.15 |
ENSDART00000076915
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr3_+_15586954 | 0.15 |
ENSDART00000080075
|
phb
|
prohibitin |
| chr5_-_33156615 | 0.15 |
ENSDART00000159058
|
dab2ipb
|
DAB2 interacting protein b |
| chr17_-_31594647 | 0.15 |
ENSDART00000135468
|
si:dkey-170l10.1
|
si:dkey-170l10.1 |
| chr5_-_27958815 | 0.14 |
ENSDART00000177313
|
si:ch73-195i19.3
|
si:ch73-195i19.3 |
| chr11_-_25181234 | 0.14 |
ENSDART00000013714
|
gata1a
|
GATA binding protein 1a |
| chr20_-_16553010 | 0.14 |
|
|
|
| chr5_+_36431824 | 0.14 |
ENSDART00000045036
|
nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr13_+_32609784 | 0.13 |
ENSDART00000160138
|
sobpa
|
sine oculis binding protein homolog (Drosophila) a |
| chr4_-_19992951 | 0.13 |
ENSDART00000169248
|
drd4-rs
|
dopamine receptor D4 related sequence |
| chr9_+_4708975 | 0.13 |
ENSDART00000081326
|
prpf40a
|
PRP40 pre-mRNA processing factor 40 homolog A |
| chr8_+_23334921 | 0.13 |
ENSDART00000085361
ENSDART00000046460 ENSDART00000145062 |
dnmt3ba
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate a |
| chr8_-_23591082 | 0.13 |
ENSDART00000025024
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr8_-_46903869 | 0.13 |
ENSDART00000161462
|
acot7
|
acyl-CoA thioesterase 7 |
| chr4_+_14658179 | 0.13 |
ENSDART00000168152
|
abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr17_+_25425967 | 0.12 |
ENSDART00000030691
|
clic4
|
chloride intracellular channel 4 |
| chr13_-_40027211 | 0.12 |
|
|
|
| chr2_+_50873893 | 0.12 |
|
|
|
| chr23_-_36319185 | 0.11 |
ENSDART00000139328
|
znf740b
|
zinc finger protein 740b |
| chr2_-_26160882 | 0.11 |
ENSDART00000133163
|
CR392026.2
|
ENSDARG00000094803 |
| chr6_+_33896684 | 0.11 |
ENSDART00000165710
|
gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr7_-_25624128 | 0.11 |
ENSDART00000173505
|
cd99l2
|
CD99 molecule-like 2 |
| chr5_+_65754237 | 0.11 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr3_-_38642067 | 0.10 |
ENSDART00000155518
|
znf281a
|
zinc finger protein 281a |
| chr11_+_24582928 | 0.09 |
ENSDART00000135443
|
kdm5ba
|
lysine (K)-specific demethylase 5Ba |
| chr6_+_3120600 | 0.09 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr9_-_39190534 | 0.09 |
|
|
|
| chr6_-_3837266 | 0.08 |
ENSDART00000059212
|
unc50
|
unc-50 homolog (C. elegans) |
| chr8_+_39964882 | 0.08 |
ENSDART00000134452
|
ggt5a
|
gamma-glutamyltransferase 5a |
| chr10_+_37193735 | 0.08 |
ENSDART00000114909
|
cuedc1a
|
CUE domain containing 1a |
| chr14_+_4169371 | 0.08 |
ENSDART00000136665
|
gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr2_-_6545431 | 0.07 |
ENSDART00000161934
|
si:dkey-119f1.1
|
si:dkey-119f1.1 |
| chr20_-_15029379 | 0.07 |
ENSDART00000152641
|
dnm3a
|
dynamin 3a |
| chr13_+_32609849 | 0.07 |
ENSDART00000160138
|
sobpa
|
sine oculis binding protein homolog (Drosophila) a |
| chr16_-_16212890 | 0.07 |
ENSDART00000131227
|
ankib1b
|
ankyrin repeat and IBR domain containing 1b |
| chr25_-_27120954 | 0.07 |
ENSDART00000157319
|
hyal4
|
hyaluronoglucosaminidase 4 |
| chr10_+_3461461 | 0.06 |
|
|
|
| chr19_+_7019317 | 0.06 |
ENSDART00000139122
|
flot1a
|
flotillin 1a |
| chr1_+_9634016 | 0.06 |
ENSDART00000029774
|
tmem55bb
|
transmembrane protein 55Bb |
| chr8_+_25015374 | 0.06 |
ENSDART00000140617
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr1_-_8439215 | 0.06 |
ENSDART00000081337
|
ndufab1a
|
NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1a |
| chr25_-_20160885 | 0.06 |
ENSDART00000160700
|
dnm1l
|
dynamin 1-like |
| chr20_+_31173383 | 0.05 |
ENSDART00000136255
|
otofa
|
otoferlin a |
| chr8_-_8408046 | 0.05 |
ENSDART00000132700
|
cdk16
|
cyclin-dependent kinase 16 |
| chr11_-_18081944 | 0.05 |
ENSDART00000113468
|
CABZ01112215.1
|
ENSDARG00000079534 |
| chr19_-_19806070 | 0.05 |
ENSDART00000165206
|
smarcc1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1b |
| chr9_-_14302522 | 0.04 |
ENSDART00000135458
|
abcb6b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6b |
| chr21_-_11561793 | 0.04 |
ENSDART00000171708
ENSDART00000081614 |
cast
|
calpastatin |
| chr3_+_27655753 | 0.04 |
ENSDART00000086994
|
nat15
|
N-acetyltransferase 15 (GCN5-related, putative) |
| chr2_-_25306278 | 0.04 |
ENSDART00000132050
|
hltf
|
helicase-like transcription factor |
| chr24_+_16402613 | 0.03 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr21_-_44707326 | 0.03 |
ENSDART00000013814
|
ammecr1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 homolog (human) |
| chr2_+_50873843 | 0.03 |
|
|
|
| chr12_-_33871785 | 0.03 |
ENSDART00000105545
|
arl3
|
ADP-ribosylation factor-like 3 |
| chr11_+_35790782 | 0.03 |
ENSDART00000125221
|
CR933559.1
|
ENSDARG00000087939 |
| chr13_+_36496475 | 0.03 |
ENSDART00000133198
|
si:ch211-67f24.7
|
si:ch211-67f24.7 |
| chr10_-_8094671 | 0.02 |
ENSDART00000099033
|
zgc:158494
|
zgc:158494 |
| chr23_+_28882409 | 0.02 |
ENSDART00000078171
|
pex14
|
peroxisomal biogenesis factor 14 |
| chr3_+_32700750 | 0.02 |
ENSDART00000139410
|
cd2bp2
|
CD2 (cytoplasmic tail) binding protein 2 |
| chr3_-_16569378 | 0.02 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr25_-_3345176 | 0.02 |
ENSDART00000029067
|
hbp1
|
HMG-box transcription factor 1 |
| chr3_-_13311188 | 0.02 |
ENSDART00000080807
|
fbxw9
|
F-box and WD repeat domain containing 9 |
| chr10_+_29964164 | 0.02 |
ENSDART00000118838
|
SNORD14
|
Small nucleolar RNA SNORD14 |
| chr7_+_24787720 | 0.01 |
|
|
|
| chr16_-_42105733 | 0.01 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr10_-_28142344 | 0.01 |
ENSDART00000023545
|
ints2
|
integrator complex subunit 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 | 0.4 | GO:0052803 | imidazole-containing compound metabolic process(GO:0052803) imidazole-containing compound catabolic process(GO:0052805) |
| 0.1 | 0.4 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.1 | 0.5 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 0.5 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.1 | 0.9 | GO:0003315 | heart rudiment formation(GO:0003315) |
| 0.1 | 0.4 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 0.2 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.1 | 0.6 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 1.2 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.1 | 0.4 | GO:1901099 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.4 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.1 | 0.4 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 0.6 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.1 | 0.6 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 0.2 | GO:0014829 | phasic smooth muscle contraction(GO:0014821) vascular smooth muscle contraction(GO:0014829) endocrine process(GO:0050886) |
| 0.1 | 0.2 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.1 | 1.5 | GO:0071698 | olfactory placode development(GO:0071698) |
| 0.1 | 0.4 | GO:0070571 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.6 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.0 | 0.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.2 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:0031280 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclic nucleotide metabolic process(GO:0030800) negative regulation of cyclic nucleotide biosynthetic process(GO:0030803) negative regulation of nucleotide biosynthetic process(GO:0030809) negative regulation of cAMP metabolic process(GO:0030815) negative regulation of cAMP biosynthetic process(GO:0030818) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) negative regulation of purine nucleotide biosynthetic process(GO:1900372) |
| 0.0 | 0.2 | GO:0015859 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.0 | 0.2 | GO:0046098 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine metabolic process(GO:0046098) guanine biosynthetic process(GO:0046099) hypoxanthine metabolic process(GO:0046100) |
| 0.0 | 0.2 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.1 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.4 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.0 | 0.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.3 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.2 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.2 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.4 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.4 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.6 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.1 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.3 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.3 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 0.1 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.1 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.1 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.1 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.1 | 0.4 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 0.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.5 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.5 | GO:0005119 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.4 | GO:0016841 | folic acid binding(GO:0005542) ammonia-lyase activity(GO:0016841) |
| 0.1 | 0.5 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 0.3 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.3 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.4 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.1 | 0.3 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.4 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.2 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.6 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 1.9 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.1 | GO:0099528 | G-protein coupled neurotransmitter receptor activity(GO:0099528) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.9 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.1 | GO:0051192 | ACP phosphopantetheine attachment site binding involved in fatty acid biosynthetic process(GO:0000036) ACP phosphopantetheine attachment site binding(GO:0044620) prosthetic group binding(GO:0051192) |
| 0.0 | 0.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.4 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 1.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.7 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.3 | REACTOME BILE ACID AND BILE SALT METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.0 | 0.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.1 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 1.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.1 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.7 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |